Introduction
With the advent of RAG (Retrieval Augmented Generation) and Large Language Models (LLMs), knowledge-intensive tasks like Document Question Answering, have become a lot more efficient and robust without the immediate need to fine-tune a cost-expensive LLM to solve downstream tasks. In this article, we will dive into the world of RAG-powered document QnA using Google’s Gemini AI and Langchain. Alongside this, there has been a lot of discussion around preserving conversational memory while leveraging LLMs for QnA. With that in mind, we will also learn how to create a custom semantic memory and integrate it with our RAG to aid a conversational interface where the user can ask follow-up questions and keep the chat going. With that, let’s dig in!
Learning Objectives
- Read and store PDF documents in a vector store using Gemini Embeddings.
- Create a custom PDF reader to insert metadata information based on our choice and use case.
- Generate responses for user queries with the help of the Gemini Pro model.
- Implement semantic caching to store LLM responses to queries and use them as similar query responses.
This article was published as a part of the Data Science Blogathon.
Table of Contents
Conversing with Documents
Creating a Document Question Answering application is much easier now than it was a year ago. OpenAI API has been the core choice for most of the RAG applications since its launch, but for small applications with less/no funding, OpenAI becomes an expensive choice. This is where Google’s Gemini API catches attention. The free version of the API supports up to 60 QPM, which might seem less but is helpful for non-customer-centric applications or for hobbyists who don’t want to spend dollars on their projects.
Additionally, in this article, we will also be implementing semantic caching, which will be helpful for applications that are using the paid version of OpenAI API. Now, before we jump into the hands-on actions, let’s understand what semantic caching is.
What is Semantic Caching?
Semantic caching is a kind of caching that can be used to cache LLM responses. It stores LLM response for query and returns the same response when the same query or similar query is asked. This helps reduce unnecessary LLM calls and consequently reduces API costs.
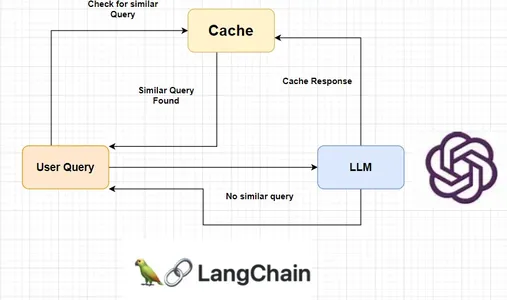
Langchain provides a wide range of LLM cache tools like Redis Semantic Cache, GPTCache, AstraDB, etc. However, they are not accurate in terms of accurately recognizing similar queries. This happens due to high similarity scores for queries that differ by a number or word. Well, we will also handle the issue by creating our own semantic cache system using the same concept.
Hands-On Document QnA with Langchain + Gemini Pro with Semantic Caching
The first step in building this RAG system is building the Ingestion pipeline. The Ingestion pipeline simply allows the user to upload the document(s) for question answering. We initialize a vector store and then store the document contents in it. We will store document contents, their embeddings, and document metadata information in the vector store.
For the embeddings, we will be using the embeddings model provided by Gemini. We will also create our PDF reader to add extra information like page numbers, file names, etc., within the metadata. We will use the Deeplake vector store for storing the documents for RAG as well as for semantic caching later in the QA chain.
Step-by-Step Guide to Document QnA with Langchain + Gemini Pro
Let’s begin the process of creating a document QnA system using Google Gemini Pro and Langchain, with this detailed 4-step guide.
Step 1: Creating Gemini API Key
The first step will be creating an API key for Gemini AI. You can skip this step if you already have a key ready. Else, follow the below steps to create a new API key:
- Go to https://aistudio.google.com/app
- Click on Get API key -> Create API key.
- Click on the `Create API key on a new project` or `Search Google Cloud Projects` and select a project. Then wait for the key to be generated.
- Copy the generated API key.
Now put this key in the config.py file in the GOOGLE_API_KEY variable. We are now good to go. Create a virtual environment using the tool of your choice and use the requirements.txt file provided to install the required packages. Please avoid changing the provided version of the packages as that might break your final application.
Here are the contents of the requirements.txt file:
deeplake==3.8.19
langchain==0.1.3
langchain-community==0.0.20
langchain-core==0.1.23
langchain-google-genai==0.0.11
langsmith==0.0.87
lxml==4.9.4
nltk==3.8.1
numpy==1.26.2
openai==0.28.0
pydantic==2.5.3
pydantic_core==2.14.6
PyMuPDF==1.23.21
pypdf==3.17.4
pypdfium2==4.25.0
scipy==1.12.0
sentence-transformers==2.3.1
tiktoken==0.5.2
transformers==4.36.2Run the following code to install all the necessary packages:
# For linux and macOS systems:
pip install -r requirements.txt
# For Windows systems:
python -m pip install -r requirements.txtBelow is the entire codebase for the Ingestion pipeline:
import config as cfg
from langchain.vectorstores.deeplake import DeepLake
from src.pdf_reader import PDFReader
from langchain_google_genai import (
GoogleGenerativeAIEmbeddings,
)
class Ingestion:
"""Ingestion class for ingesting documents to vectorstore."""
def __init__(self):
self.text_vectorstore = None
self.image_vectorstore = None
self.text_retriever = None
self.embeddings = GoogleGenerativeAIEmbeddings(
model="models/embedding-001",
google_api_key=cfg.GOOGLE_API_KEY,
)
def ingest_documents(
self,
file: str,
):
# Initialize the PDFReader and load the PDF as chunks
loader = PDFReader()
chunks = loader.load_pdf(file_path=file)
# Initialize the vector store
vstore = DeepLake(
dataset_path="database/text_vectorstore",
embedding=self.embeddings,
overwrite=True,
num_workers=4,
verbose=False,
)
# Ingest the chunks
_ = vstore.add_documents(chunks)Let’s break down the code and understand each line quickly. We have an Ingestion class that can be initialized. In its constructor, we have initialized the Gemini Embeddings using the model name and API key, which we import from the config file. It is noteworthy that, we can use any embeddings model here and not limited to the usage of Gemini Embeddings.
We then create a ingest_documents method, that reads the documents and dumps the content into the vector store. We do that by initializing our custom PDF reader and then splitting the document into chunks. This is handled internally by our PDF reader, which will be discussed in the following section.
Now that we have document chunks we can now ingest them into the vector database. We initialize the Deeplake vector store using the path and the embeddings we initialized earlier in the constructor. Additionally, we set the overwrite parameter to True so that the previous contents in the vector store are overwritten. We then add the extracted chunks from the document to the vector store.
Step 2: Reading, Loading, and Processing the PDF
The PDF reader that we initialized earlier is a custom PDF reader that we will create for our use case. We will make use of Langchain’s PyPDFLoader to load the PDF document and CharacterTextSplitter to split the document into chunks of smaller size. Then we update the chunks’ metadata using our preferred information like page number and filename. Below is the entire codebase for the PDF Reader tool:
import os
import config as cfg
from langchain.document_loaders.pdf import PyPDFLoader
from langchain.text_splitter import (
CharacterTextSplitter,
)
from langchain.schema import Document
class PDFReader:
"""Custom PDF Loader to embed metadata with the pdfs."""
def __init__(self) -> None:
self.file_name = ""
self.total_pages = 0
def load_pdf(self, file_path):
# Get the filename from file path
self.file_name = os.path.basename(file_path)
# Initialize Langchain's PyPDFLoader to load the PDF pages
loader = PyPDFLoader(file_path)
# Initialize the text splitter
text_splitter = CharacterTextSplitter(
separator="\n",
chunk_size=cfg.PDF_CHARSPLITTER_CHUNKSIZE,
chunk_overlap=cfg.PDF_CHARSPLITTER_CHUNK_OVERLAP,
)
# Load the pages from the document
pages = loader.load()
self.total_pages = len(pages)
chunks = []
# Loop through the pages
for idx, page in enumerate(pages):
# Append each page as Document object with modified metadata
chunks.append(
Document(
page_content=page.page_content,
metadata=dict(
{
"file_name": self.file_name,
"page_no": str(idx + 1),
"total_pages": str(self.total_pages),
}
),
)
)
# Split the documents using splitter
final_chunks = text_splitter.split_documents(chunks)
return final_chunksLet’s break down the code and understand each line quickly. The class constructor has two local variables file_name and total_pages which are initialized as empty string and 0 respectively. The class has a main method called load_pdf which loads the document contents and splits them into smaller chunks. We first initialize Langchain’s PyPDFLoader using the file path. Then initialize the Character splitter object which will be used later to split the chunks. The Character splitter takes in 3 arguments: separator, chunk size, and chunk overlap.
The default separator is ‘\n\n’ which is not helpful while using chunk overlap, so we set it to ‘\n’. We can have chunk size and chunk overlap of our choice. For this example, we can set them to 1000 and 200 respectively. Then we load the document using the loader instance and get the total number of pages.
We then iterate over the loaded pages from PDF and create a new list of document objects using the page_content and our metadata information. At this stage, we have a list of chunks of the document with additional metadata. Then we proceed to split the chunks using the text splitter and return them.
Step 3: Building the Semantic Cache
Next, let’s build our semantic cache service for storing LLM responses and using them as responses for similar queries. Below is the code for the CustomGPTCache tool:
from typing import List
import config as cfg
from langchain.schema import Document
from langchain.vectorstores.deeplake import DeepLake
from langchain.embeddings.sentence_transformer import SentenceTransformerEmbeddings
class CustomGPTCache:
def __init__(self) -> None:
# Initialize the embeddings model and cache vector store
self.embeddings = SentenceTransformerEmbeddings(
model_name="all-MiniLM-L12-v2"
)
self.response_cache_store = DeepLake(
dataset_path="database/cache_vectorstore",
embedding=self.embeddings,
read_only=False,
num_workers=4,
verbose=False,
)
def cache_query_response(self, query: str, response: str):
# Create a Document object using query as the content and it's
# response as metadata
doc = Document(
page_content=query,
metadata={"response": response},
)
# Insert the Document object into cache vectorstore
_ = self.response_cache_store.add_documents(documents=[doc])
def find_similar_query_response(self, query: str, threshold: int):
try:
# Find similar query based on the input query
sim_response = self.response_cache_store.similarity_search_with_score(
query=query, k=1
)
# Return the response from the fetched entry if it's score is more
# than threshold
return [
{
"response": res[0].metadata["response"],
}
for res in sim_response
if res[1] > threshold
]
except Exception as e:
raise Exception(e)Let’s walk through how we are building the cache system. We are using the all-MiniLM-L12-v2 model from Sentence Transformers for creating embeddings for the cache. The motivation for this approach was that: for embeddings of higher dimension, we get a higher range of similarity score between dissimilar queries which results in mis-triggering of the cache.
From extensive research, I have observed that using embeddings of lower dimensions, say 256, we get a wider range of similarity scores between 0.3 to 1.0. Coming back to the code, we initialize the embeddings using Langchain’s SentenceTransformerEmbeddings by specifying the model name. Next, we initialize the Deeplake vector store where the query and responses will be stored.
Note that when initializing cache vector store we don’t set the overwrite parameter to True as that would be counter-intuitive. We will add two methods to the CustomGPTCache class: cache_query_response and find_similar_query_response. Let’s discuss about them.
- cache_query_response: This method is called in the QAChain when an LLM response is generated. A Document object is created with the query in the page_content and the response in the metadata field. Then, that Document object is added to the vector store.
- find_similar_query_response: This method is called in the QAChain for every run. It returns a list of dictionaries containing responses. The user query is passed into this method for every run. It uses the query to do a similarity search on the cache vector store to find the most similar query that is cached. We only fetch one similar entry from the vector store. Then we create a list of those responses and put a threshold on the similarity score to prevent returning irrelevant responses.
Step 4: Document Questions and Answering Using QAChain
Finally, we will look at how QAChain utilizes the cache vector store and docstore to generate responses to user queries. We initialize Gemini embeddings and Gemini model in the class constructor using the necessary parameters like model name, API key, etc. We also initialize the cache here which will be used in the other methods. Then, we will add two methods to the QAChain class for handling user queries: generate_response and ask_question.
- generate_response: This method handles the LLM response part of the QAChain. It simply takes a user query and generates a response for the query using Langchain’s RetrievalQA Chain. After the response is generated, it is cached using the cache_query_response method of CustomGPTCache, and the response is returned.
- 2. ask_question: This method is called for every user response. First, the find_similar_query_response method of CustomGPTCache is called using the user query. If it returns a list of responses, the response is returned, otherwise, the generate_response method is called using the user query. Additionally, the generate_response method is called if any exception is raised from the cache side. Below is the codebase for the QAChain pipeline for reference.
import config as cfg
from src.cache import CustomGPTCache
from langchain.chains import RetrievalQA
from langchain.prompts import PromptTemplate
from langchain.vectorstores.deeplake import DeepLake
from langchain_google_genai import (
GoogleGenerativeAIEmbeddings,
ChatGoogleGenerativeAI
)
class QAChain:
def __init__(self) -> None:
# Initialize Gemini Embeddings
self.embeddings = GoogleGenerativeAIEmbeddings(
model="models/embedding-001",
google_api_key=cfg.GOOGLE_API_KEY,
task_type="retrieval_query",
)
# Initialize Gemini Chat model
self.model = ChatGoogleGenerativeAI(
model="gemini-pro",
temperature=0.3,
google_api_key=cfg.GOOGLE_API_KEY,
convert_system_message_to_human=True,
)
# Initialize GPT Cache
self.cache = CustomGPTCache()
self.text_vectorstore = None
self.text_retriever = None
def ask_question(self, query):
try:
# Search for similar query response in cache
cached_response = self.cache.find_similar_query_response(
query=query, threshold=cfg.CACHE_THRESHOLD
)
# If similar query response is present,vreturn it
if len(cached_response) > 0:
print("Using cache")
result = cached_response[0]["response"]
# Else generate response for the query
else:
print("Generating response")
result = self.generate_response(query=query)
except Exception as _:
print("Exception raised. Generating response.")
result = self.generate_response(query=query)
return result
def generate_response(self, query: str):
# Initialize the vectorstore and retriever object
vstore = DeepLake(
dataset_path="database/text_vectorstore",
embedding=self.embeddings,
read_only=True,
num_workers=4,
verbose=False,
)
retriever = vstore.as_retriever(search_type="similarity")
retriever.search_kwargs["distance_metric"] = "cos"
retriever.search_kwargs["fetch_k"] = 20
retriever.search_kwargs["k"] = 15
# Write prompt to guide the LLM to generate response
prompt_template = """
<YOUR PROMPT HERE>
Context: {context}
Question: {question}
Answer:
"""
PROMPT = PromptTemplate(
template=prompt_template, input_variables=["context", "question"]
)
chain_type_kwargs = {"prompt": PROMPT}
# Create Retrieval QA chain
qa = RetrievalQA.from_chain_type(
llm=self.model,
retriever=retriever,
verbose=False,
chain_type_kwargs=chain_type_kwargs,
)
# Run the QA chain and store the response in cache
result = qa({"query": query})["result"]
self.cache.cache_query_response(query=query, response=result)
return resultNow that we have the entire pipeline ready, let’s use them to test our application. The first step is to ingest the document. We initialize the Ingestion object and call the ingest_document method using the file path for the document that is to be stored.
from src.ingestion import Ingestion
ingestion = Ingestion()
file = "Apple 10k.pdf"
ingestion.ingest_documents(
file=file
)After the document is ingested, we will initialize the QAChain object. Then we call the ask_question method using the user query to generate a response.
from src.qachain import QAChain
qna = QAChain()
%%time
query = "What were the highlights for 2nd quarter of FY2023?"
results = qna.ask_question(
query=query
)
print(results)
# OUTPUT:
# Generating response
# The highlights for the second quarter of FY2023 were:
# • MacBook Pro 14”, MacBook Pro 16” and Mac mini; and
# • Second-generation HomePod.
# CPU times: total: 2.94 s
# Wall time: 11.4 s
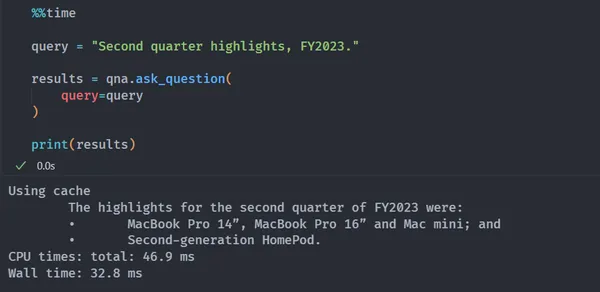
%%time
query = "Second quarter highlights, FY2023."
results = qna.ask_question(
query=query
)
print(results)
# OUTPUT:
# Using cache
# The highlights for the second quarter of FY2023 were:
# • MacBook Pro 14”, MacBook Pro 16” and Mac mini; and
# • Second-generation HomePod.
# CPU times: total: 46.9 ms
# Wall time: 32.8 ms
Application Performance and Limitations
There are several RAG evaluation libraries that can be used to evaluate the performance of the application. The two most popular ones are RAGAS and Tonic Validate Metrics. Both offer similar metrics like Answer similarity score, Retrieval precision, Augmentation precision, Augmentation accuracy/relevance, and Retrieval k-recall, which help evaluate the application performance. Although few exceptions are handled in the above codebases, there are a few cases where the application might crash. The PDFReader currently has no check on the input file type which needs to be handled.
This application is extensively reusable in a wide range of business use cases like medical, financial, industrial, e-commerce, etc. In medical industries, RAG pipelines can help in quickly troubleshooting instrument failures, deciding the correct medicine for a medical condition, etc. RAG pipelines can help answer queries from a large corpus of financial documents in the blink of an eye. Their quick retrieval and answering capabilities make them ideal for question answering over large datasets. RAG pipelines can also enable quick review of previous research advancements of a topic and suggest further research scope.
While our current pipeline is ready to be used for any use case, there are certain limitations. Gemini pro model currently has a context length of 32k which might not be suitable for large knowledge bases. An alternate suggestion would be to use the Gemini Pro 1.5 model which supports a context length of up to 1 million. The current pipeline has no chat memory, so, the model will not have the context to previous conversations. Langchain provides several memory options (Memory Docs) that can be integrated into the pipeline effortlessly.
Code Reusability
- The Ingestion pipeline code can be reused to build ingestion systems for other applications that require document storage and retrieval.
- The PDF Reader tool can be repurposed for other applications that require processing and metadata extraction from PDF documents.
- The CustomGPTCache code can be used in other projects to implement efficient caching and retrieval of LLM responses or similar use cases.
- The QAChain class can be adapted for other question-answering systems that use document stores and caching mechanisms.
Conclusion
So, folks, that’s how you can communicate with your documents through RAG-powered question answering and semantic caching using Gemini Pro and Langchain!
In this article, we’ve developed a pipeline that showcases a comprehensive approach to handling documents for intelligent QnA. By integrating Google’s Gemini embeddings, a custom PDF reader, and semantic caching, this RAG system maintains efficiency and accuracy in providing answers to user queries. Moreover, the codebase is highly adaptable scalable, and reliable for a variety of QnA use cases with minimal code changes!
Key Takeaways
- The RAG system involves building an Ingestion pipeline to upload and store documents for question-answering.
- The Ingestion pipeline uses Gemini embeddings for document embeddings and a custom PDF reader for metadata extraction.
- A custom semantic cache service is implemented to store and retrieve LLM responses for similar queries efficiently.
- The QAChain class is responsible for generating responses to user queries and utilizing the cache and vector store.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.





