Introduction
In the previous article, we experimented with Cohere’s Command-R model and Rerank model to generate responses and rerank doc sources. We have implemented a simple RAG pipeline using them to generate responses to user’s questions on ingested documents. However, what we have implemented is very simple and unsuitable for the general user, as it has no user interface to interact with the chatbot directly. In this article, we will modularize the codebase for easy interpretation and scaling and build a Streamlit application that will serve as an interface to interact with the RAG pipeline. The interface will be a chatbot interface that the user can use to interact with it. So, we will implement an additional memory component within the application, allowing users to ask follow-up queries on previous responses.

Learning Objectives
- Using object-oriented programming (OOP) principles, develop a reusable, modular codebase for various RAG pipelines.
- Create an ingestion pipeline for document ingestion components and a query pipeline for query-related components. Both are independent and can run separately.
- Connect only the query pipeline to the Streamlit app for user queries, with an option to add document ingestion by modifying the code.
- Implement a memory component to enable follow-up queries based on previous responses.
- Turn notebook experiments into demo-able applications within the Python ecosystem.
- Facilitate faster prototype development with minimal code changes by creating reusable code for future RAG pipelines.
Table of contents
This article was published as a part of the Data Science Blogathon.
Document QnA Pipeline Development
The first step in building a prototype or deployable application is defining the configurations and constants used within various application sections. The application has several configurable options, such as chunk size and overlap in the Ingestion pipeline, the API key for Cohere endpoints, and the temperature for LLM generation. These configurations will be in a central config file, accessible from anywhere within the application.
We will need to follow a folder structure for this project. We will have a ‘src’ directory where all the necessary files will be stored, and the app.py file will be in the root directory. Below is the structure that we will follow:
.
├── .venv
├── src
│ ├── config.py
│ ├── constants.py
│ ├── ingestion.py
│ └── qna.py
├── app.py
└── requirements.txtWe will create two files for two purposes: A config.py file to hold the secret keys, a vector store path, and a few other configurations and a constants.py file to hold all the constants used in the application like the chunk size, chunk overlap, and prompt template. Below are the contents for the config.py file:
COHERE_EMBEDDING_MODEL_NAME = "embed-english-v3.0"
COHERE_MODEL_NAME = "command-r"
COHERE_RERANK_MODEL_NAME = "rerank-english-v3.0"
DEEPLAKE_VECTORSTORE = "/path/to/document/vectorstore"
API_KEY = “”
Below are the contents for constants.py file:
PDF_CHARSPLITTER_CHUNKSIZE = 1000
PDF_CHARSPLITTER_CHUNK_OVERLAP = 100
TEMPERATURE = 0.3
TOP_K = 25
CONTEXT_THRESHOLD = 0.8
PROMPT_TEMPLATE = """
<YOUR PROMPT HERE>
Chat History: {chat_history} Context: {context} Question: {question} Answer:
"""In the config.py file, I have put the Cohere API key, names of all the models used, and path to the document vector store. In the constants.py file, I have put prompt template and other ingestion and generation configurations like chunk size and chunk overlap values, temperature for LLM generation, top_k for the topmost relevant chunks, and the context threshold to filter out chunks that have relevancy
score below 0.8. The contents of the config.py and constants.py files can be changed based on use cases.
Part 1 – Ingestion
Next, we will look at how we can modularize the Ingestion pipeline. We will create a single class named Ingestion and add a method to generate embeddings and store them in the vector store. Note
that we will have single files for our use case for each pipeline. As the complexity of the use case increases, multiple files can be created to handle each pipeline component. This will ensure
code readability and ease in further changes and updates.
Below is the code for the Ingestion class:
import timeimport time
import src.constants as constant
import src.config as cfg
from langchain_cohere import CohereEmbeddings
from langchain.document_loaders.pdf import PyPDFLoader
from langchain.text_splitter import CharacterTextSplitter
class Ingestion:
def __init__(self):
self.text_vectorstore = None
self.embeddings = CohereEmbeddings(
model=cfg.COHERE_EMBEDDING_MODEL_NAME,
cohere_api_key=cfg.API_KEY,
)
def create_and_add_embeddings(
self,
file_path: str,
):
self.text_vectorstore = DeepLake(
dataset_path=cfg.DEEPLAKE_VECTORSTORE,
embedding=self.embeddings,
verbose=False,
num_workers=4,
)
loader = PyPDFLoader(file_path=file_path)
text_splitter = CharacterTextSplitter(
separator="\n",
chunk_size=constant.PDF_CHARSPLITTER_CHUNKSIZE,
chunk_overlap=constant.PDF_CHARSPLITTER_CHUNK_OVERLAP,
)
pages = loader.load()
chunks = text_splitter.split_documents(pages)
_ = self.text_vectorstore.add_documents(documents=chunks)
import src.constants as constant
import src.config as cfg
from langchain_cohere import CohereEmbeddings
from langchain.document_loaders.pdf import PyPDFLoader
from langchain.text_splitter import CharacterTextSplitter
class Ingestion:
def __init__(self):
self.text_vectorstore = None
self.embeddings = CohereEmbeddings(
model=cfg.COHERE_EMBEDDING_MODEL_NAME,
cohere_api_key=cfg.API_KEY,
)
def create_and_add_embeddings(
self,
file_path: str,
):
self.text_vectorstore = DeepLake(
dataset_path=cfg.DEEPLAKE_VECTORSTORE,
embedding=self.embeddings,
verbose=False,
num_workers=4,
)
loader = PyPDFLoader(file_path=file_path)
text_splitter = CharacterTextSplitter(
separator="\n",
chunk_size=constant.PDF_CHARSPLITTER_CHUNKSIZE,
chunk_overlap=constant.PDF_CHARSPLITTER_CHUNK_OVERLAP,
)
pages = loader.load()
chunks = text_splitter.split_documents(pages)
_ = self.text_vectorstore.add_documents(documents=chunks)Let’s understand each part of the above code. First, we import all necessary packages, including the constants and config files. Then, we define the class Ingestion and its class constructor using the __init__ method. We set the text_vectorstore variable to None, which will be initialized with the vector store instance later. Then, we initialize the Embeddings model instance using the model name and the API key from the config.
Next, we create the create_and_add_embeddings method, which takes the file_path to which the document is ingested. Inside this method, we first initialize the vector store using the vector store path and embeddings. We have also set the num_workers to 4 so that 4 CPU cores are utilized for faster processing. Then, we initialize the PDF Loader object using the file_path, and then we use the Character Splitter to split the chunks. We then load the PDF file and split the pages into further chunks. The final chunks are then added to the vector store.
Part 2 – QnA
Now that we have the ingestion pipeline setup, we will create the QnA pipeline. Below is the code for the QnA class:
import time
import src.constants as constant
import src.config as cfg
from pymongo import MongoClient
from langchain_cohere import CohereEmbeddings
from langchain_cohere import ChatCohere
from langchain.memory.chat_message_histories.sql import SQLChatMessageHistory
from langchain.memory import ConversationBufferWindowMemory
from langchain.chains.conversational_retrieval.base import ConversationalRetrievalChain
from langchain.prompts import PromptTemplate
from langchain.retrievers import ContextualCompressionRetriever
from langchain_cohere import CohereRerank
class QnA:
def __init__(self):
self.embeddings = CohereEmbeddings(
model=cfg.COHERE_EMBEDDING_MODEL_NAME,
cohere_api_key=cfg.API_KEY,
)
self.model = ChatCohere(
model=cfg.COHERE_MODEL_NAME,
cohere_api_key=cfg.API_KEY,
temperature=constant.TEMPERATURE,
)
self.cohere_rerank = CohereRerank(
cohere_api_key=cfg.API_KEY,
model=cfg.COHERE_RERANK_MODEL_NAME,
)
self.text_vectorstore = None
self.text_retriever = None
def ask_question(
self,
query,
session_id,
verbose: bool = False,
):
start_time = time.time()
self.init_vectorstore()
memory_key = "chat_history"
history = SQLChatMessageHistory(
session_id=session_id,
connection_string="sqlite:///memory.db",
)
PROMPT = PromptTemplate(
template=constant.PROMPT_TEMPLATE,
input_variables=["chat_history", "context", "question"],
)
memory = ConversationBufferWindowMemory(
memory_key=memory_key,
output_key="answer",
input_key="question",
chat_memory=history,
k=2,
return_messages=True,
)
chain_type_kwargs = {"prompt": PROMPT}
qa = ConversationalRetrievalChain.from_llm(
llm=self.model,
combine_docs_chain_kwargs=chain_type_kwargs,
retriever=self.text_retriever,
verbose=verbose,
memory=memory,
return_source_documents=True,
chain_type="stuff",
)
response = qa.invoke({"question": query})
exec_time = time.time() - start_time
return response
def init_vectorstore(self):
self.text_vectorstore = DeepLake(
dataset_path=cfg.DEEPLAKE_VECTORSTORE,
embedding=self.embeddings,
verbose=False,
read_only=True,
num_workers=4,
)
self.text_retriever = ContextualCompressionRetriever(
base_compressor=self.cohere_rerank,
base_retriever=self.text_vectorstore.as_retriever(
search_type="similarity",
search_kwargs={
"fetch_k": 20,
"k": constant.TOP_K,
},
),
)We created a QnA class with an initializer that sets up the question-answering system. It creates an instance of the CohereEmbeddings class for generating text embeddings using the model’s name and API key. It also initializes the ChatCohere class for conversational tasks with a temperature value for text randomness and the CohereRerank class for reranking responses based on relevance.
The ask_question method takes a query, session ID, and optional verbose flag. The init_vectorstore method initializes the vector database and retriever components. A memory key and an instance of SQLChatMessageHistory manages conversation history. The PromptTemplate formats the query and history, and the ConversationBufferWindowMemory manages the conversation buffer memory.
The ConversationalRetrievalChain class combines the retriever and language model for question-answering. It’s initialized with the language model, prompt template, retriever, and other settings. The invoke method generates a response based on the query and history and calculates the execution time of ask_question.
The init_vectorstore method sets up the vector database and retriever. The DeepLake instance initializes the vector database with the path, embedding model, and other parameters. The ContextualCompressionRetriever manages the retriever component with the reranking model and vector database, specifying the search type and parameters.
Part 3 – Streamlit UI
Now that both the Ingestion and QnA pipelines are ready, we will build the Streamlit interface that will utilize the pipelines. Below is the entire code for the Streamlit interface:
import streamlit as st
from src.qna import QnA
from dataclasses import dataclass
@dataclass
class Message:
actor: str
payload: str
def main():
st.set_page_config(
page_title="KnowledgeGPT",
page_icon="📖",
layout="centered",
initial_sidebar_state="collapsed",
)
st.header("📖KnowledgeGPT")
USER = "user"
ASSISTANT = "ai"
MESSAGES = "messages"
with st.spinner(text="Initializing..."):
st.session_state["qna"] = QnA()
qna = st.session_state["qna"]
if MESSAGES not in st.session_state:
st.session_state[MESSAGES] = [
Message(
actor=ASSISTANT,
payload="Hi! How can I help you?",
)
]
msg: Message
for msg in st.session_state[MESSAGES]:
st.chat_message(msg.actor).write(msg.payload)
prompt: str = st.chat_input("Enter a prompt here")
if prompt:
st.session_state[MESSAGES].append(Message(actor=USER, payload=prompt))
st.chat_message(USER).write(prompt)
with st.spinner(text="Thinking..."):
response = qna.ask_question(
query=prompt, session_id="AWDAA-adawd-ADAFAEF"
)
st.session_state[MESSAGES].append(Message(actor=ASSISTANT, payload=response))
st.chat_message(ASSISTANT).write(response)
if __name__ == "__main__":
main()Streamlit UI Functionality
The Streamlit UI serves as the user-facing component of our application. Here’s a breakdown of its functionality:
- Page Configuration: The st.set_page_config function sets the page title, icon, layout, and initial state of the sidebar.
- Constants: We define constants for the user (USER), assistant (ASSISTANT), and messages (MESSAGES) to improve code readability.
- QnA Instance Initialization: We initialize the QnA instance and store it in the st.session_state dictionary. This ensures that the instance persists across different app sessions.
- Chat Messages Initialization: If MESSAGES is not present in st.session_state, we initialize it with a welcome message from the assistant.
- Display Chat Messages: The code iterates through the MESSAGES list and displays each message along with the sender (user or assistant).
- User Input: Prompt the user to enter a prompt using st.chat_input.
- Processing User Input: If the user provides a prompt, code appends it to the MESSAGES list and generates the assistant’s response using the ask_question method of the QnA instance.
- Display Assistant Response: Append the assistant’s response to the MESSAGES list and display it to the user.
Finally, we run the main method to launch the app. We can start the app using the following command:
streamlit run app.pyWorking of the App
Below is a short demo of how the app works:
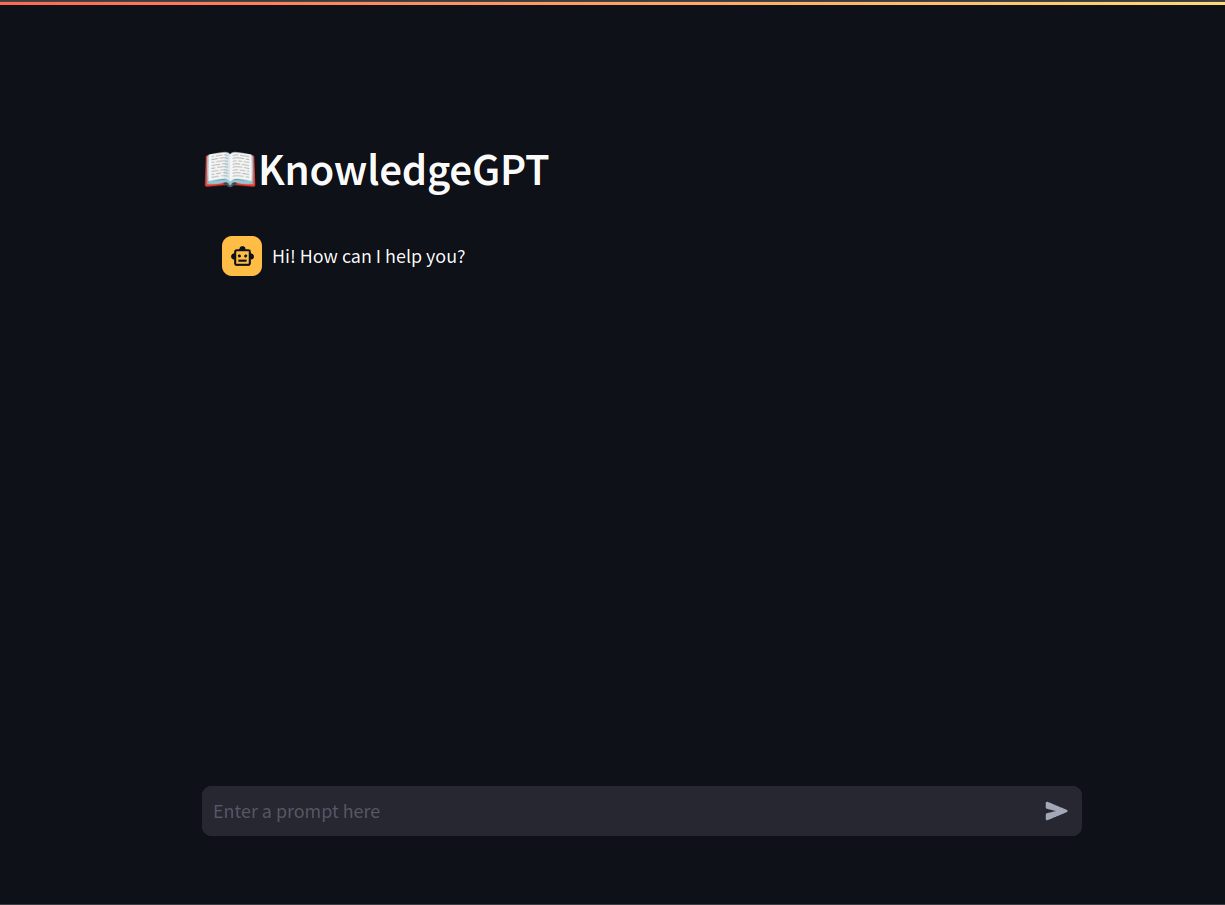
Here’s how KnowledgeGPT will work:
Conclusion
In this article, we’ve transformed our initial RAG pipeline experiment into a more robust and user-friendly application. Modifying the codebase has improved readability, maintainability, and scalability. Separate ingestion and query pipelines allow independent development and maintenance, enhancing the application’s overall scalability.
Integrating a modular backend with a Streamlit interface creates a seamless user experience through a chatbot interface that supports follow-up queries, making interactions dynamic and conversational. Using object-oriented programming principles, we’ve structured our code for clarity and reusability, which is essential for scaling and adapting to new requirements.
Our implementation of configurations and constants management, along with the setup of ingestion and QnA pipelines, provides a clear path for developers. This setup simplifies the transition from a Jupyter Notebook experiment to a deployable application, keeping the project within the Python ecosystem.
This article offers a comprehensive guide to creating an interactive document QnA application with Cohere’s models. By uniting theoretical experimentation and practical implementation, it enables developers to build efficient and scalable solutions. With the given code and clear instructions, you are now ready to develop, customize, and launch your own RAG-based applications, expediting the creation of intelligent document query systems.
Key Takeaways
- Enhances maintainability and scalability by separating ingestion and query pipelines.
- Provides a user-friendly chatbot interface for dynamic interactions.
- Ensures a structured, reusable, and scalable codebase.
- Centralized configurations in dedicated files for flexibility and ease of management.
- Efficiently handles document ingestion and user queries using Cohere’s models.
- Enables handling of follow-up queries for coherent, context-aware interactions.
- Facilitates quick prototyping and development of other RAG pipelines.
The media shown in this article are not owned by Analytics Vidhya and is used at the Author’s discretion.
Frequently Asked Questions
A. Absolutely! In fact, that is the ideal way of creating gen AI pipelines. Once the pipelines are ready, they should be wrapped with a RESTful API to be used from the frontend.
A. The Streamlit interface provides a user-friendly chatbot interface for interacting with the RAG pipeline, making it easy for users to ask questions and receive responses.
Ans. Yes. The purpose of building a modularized pipeline is to be able to stitch it to any frontend UI, be it Streamlit, Gradio, or JavaScript-based UI frameworks.





