This intelligent healthcare system uses a small language model MiniLM-L6-V2 to better understand and analyze medical information, such as symptoms or treatment instructions. The model turns text into “embeddings,” or meaningful numbers, that capture the context of the words. By using these embeddings, the system can compare symptoms effectively and make smart recommendations for conditions or treatments that match the user’s needs. This helps improve the accuracy of health-related suggestions and allows users to discover relevant care options.
Learning Objectives
- Understand how small language models generate embeddings to represent textual medical data.
- Develop skills in building a symptom-based recommendation system for healthcare applications.
- Learn methods for data manipulation and analysis using libraries like Pandas and Scikit-learn.
- Gain insights into embedding-based semantic similarity for condition matching.
- Address challenges in health-related AI systems like symptom ambiguity and data sensitivity.
This article was published as a part of the Data Science Blogathon.
Table of contents
Understanding Small Language Models
Small Language Models (SLMs) are neural language models that are designed to be computationally efficient, with fewer parameters and layers compared to larger, more resource-intensive models like BERT or GPT-3. SLMs aim to maintain a balance between lightweight architecture and the ability to perform specific tasks effectively, such as sentence similarity, sentiment analysis, and embedding generation, without requiring extensive computing resources.
Characteristics of Small Language Models
- Reduced Parameters and Layers: SLMs generally have fewer parameters (tens of millions compared to hundreds of millions or billions) and fewer layers e.g., 6 layers vs. 12 or more in larger models.
- Lower Computational Cost: They require less memory and processing power, making them faster and suitable for edge devices or applications with limited resources.
- Task-Specific Efficiency: While SLMs may not capture as much context as larger models, they are often fine-tuned for specific tasks, balancing efficiency with performance for tasks like text embeddings or document classification.

Introduction to Sentence Transformers
Sentence Transformers are models that turn text into fixed-size “embeddings,” which are like summaries in vector form that capture the text’s meaning. These embeddings make it fast and easy to compare texts, helping with tasks like finding similar sentences, searching documents, grouping similar items, and classifying text. Since the embeddings are quick to compute, Sentence Transformers are great for first pass searches.
Using All-MiniLM-L6-V2 in Healthcare
AllMiniLM-L6-v2 is a compact, pre-trained language model designed for efficient text embedding tasks. Developed as part of the Sentence Transformers framework, it uses Microsoft’s MiniLM (Minimally Distilled Language Model) architecture known for being lightweight and efficient compared to larger transformer models.
Here’s an overview of its features and capabilities:
- Architecture and Layers: The model consists of only 6 transformer layers hence the “L6” in its name, making it much smaller and faster than large models like BERT or GPT while still achieving high quality embeddings.
- Embedding Quality: Despite its small size, all-MiniLM-L6-v2 performs well for generating sentence embeddings, particularly in semantic similarity and clustering tasks. Version v2 improves performance on semantic tasks like question answering, information retrieval, and text classification through fine-tuning.
all-MiniLM-L6-v2 is an example of an SLM due to its lightweight design and specialized functionality:
- Compact Design: It has 6 layers and 22 million parameters, significantly smaller than BERT-base (110 million parameters) or GPT-2 (117 million parameters), making it both memory efficient and fast.
- Sentence Embeddings: Fine-tuned for tasks like semantic search and clustering, it produces dense sentence embeddings and achieves a high performance-to-size ratio.
- Optimized for Semantic Understanding: MiniLM models, despite their smaller size, perform well on sentence similarity and embedding-based applications, often matching the quality of larger models but with lower computational demand.
Because of these factors, AllMiniLM-L6-v2 effectively captures the main characteristics of an SLM: low parameter count, task-specific optimization, and efficiency on resource-constrained devices. This balance makes it well-suited for applications needing compact yet effective language models.
Implementing the Model in Code
Implementing the All-MiniLM-L6-V2 model in code brings efficient symptom analysis to healthcare applications. By generating embeddings, this model enables quick, accurate comparisons for symptom matching and diagnosis.
from sentence_transformers import SentenceTransformer
# 1. Load a pretrained Sentence Transformer model
model = SentenceTransformer("all-MiniLM-L6-v2")
# The sentences to encode
sentences = [
"The weather is lovely today.",
"It's so sunny outside!",
"He drove to the stadium.",
]
# 2. Calculate embeddings by calling model.encode()
embeddings = model.encode(sentences)
print(embeddings.shape)
# [3, 384]
# 3. Calculate the embedding similarities
similarities = model.similarity(embeddings, embeddings)
print(similarities)
# tensor([[1.0000, 0.6660, 0.1046],
# [0.6660, 1.0000, 0.1411],
# [0.1046, 0.1411, 1.0000]])Use Cases: Common applications of all-MiniLM-L6-v2 include:
- Semantic search, where the model encodes queries and documents for efficient similarity comparison. (e.g., in our healthcare NLP Project).
- Text classification and clustering, where embeddings help group similar texts.
- Recommendation systems, by identifying similar items based on embeddings
Building the Symptom-Based Diagnosis System
Building a symptom-based diagnosis system leverages embeddings to identify health conditions quickly and accurately. This setup translates user symptoms into actionable insights, enhancing healthcare accessibility.
Importing Necessary Libraries
!pip install sentence-transformers
import pandas as pd
from sentence_transformers import SentenceTransformer, util
# Load the data
df = pd.read_csv('/kaggle/input/disease-and-symptoms/Diseases_Symptoms.csv')
df.head()The code begins by importing the necessary libraries such as pandas and sentence-transformers for generating our text embeddings. The dataset which contains diseases and their associated symptoms, is loaded into a DataFrame df. The first few entries of the dataset are displayed. The link to the dataset.
The columns in the dataset are:
- Code: Unique identifier for the condition.
- Name: The name of the medical condition.
- Symptoms: Common symptoms associated with the condition.
- Treatments: Recommended treatments or therapies for management.

Initialize Sentence Transformer
# Initialize a Sentence Transformer model to generate embeddings
model = SentenceTransformer('all-MiniLM-L6-v2')
# Generate embeddings for each condition's symptoms
df['Symptom_Embedding'] = df['Symptoms'].apply(lambda x: model.encode(x))Then initialize a Sentence Transformer model ‘all-MiniLM-L6-v2’ this helps in converting the descriptions in the symptom column into vector embeddings. The next line of code involves applying the model to the ‘Symptoms’ column of the DataFrame asn storing the result in a new column ‘Symptom_Embedding’ that will store the embeddings for each disease’s symptoms.
# Function to find matching condition based on input symptoms
def find_condition_by_symptoms(input_symptoms):
# Generate embedding for the input symptoms
input_embedding = model.encode(input_symptoms)
# Calculate similarity scores with each condition
df['Similarity'] = df['Symptom_Embedding'].apply(lambda x: util.cos_sim(input_embedding, x).item())
# Find the most similar condition
best_match = df.loc[df['Similarity'].idxmax()]
return best_match['Name'], best_match['Treatments']Defining Function
Next, we define a function find_condition_by_symptoms() which takes the user’s input symptoms as an argument. It generates an embedding for the input symptoms and computes similarity scores between this embedding and the embeddings of the diseases in the dataset using cosine similarity. The model identifies the disease with the highest similarity score as the best match and stores it in the ‘Similarity’ column of the data frame. Using .idxmax(), it finds the index of this best match and assigns it to the variable best_match, then returns the corresponding ‘Name’ and ‘Treatments’ values.
# Example input
input_symptoms = "Sweating, Trembling, Fear of losing control"
condition_name, treatments = find_condition_by_symptoms(input_symptoms)
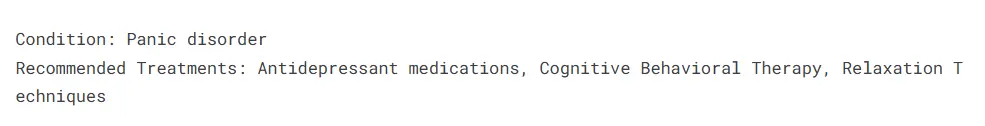
print("Condition:", condition_name)
print("Recommended Treatments:", treatments)Testing by Passing Symptoms
Finally, we provide an example input for symptoms we pass the value to find_condition_by_symptoms() function the values returned are printed, which are the name of the matching condition along with the recommended treatments. This setup allows for quick and efficient diagnosis based on user-reported symptoms.
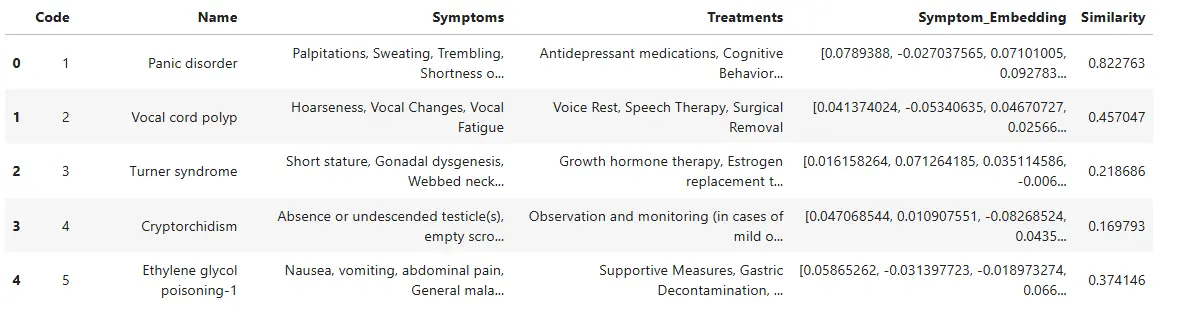
df.head()The updated data frame with the columns ‘Symptom_Embedding’ and ‘Similarity’ can be checked.
One of the challenges can be incomplete or incorrect data leading to misdiagnosis.
‘Lumps & swelling’ are common symptoms of multiple diseases for example:


Thus, the disease is highly likely misclassified by incomplete symptoms.
Challenges in Symptom Analysis and Diagnosis
Let us explore challenges in symptom analysis and diagnosis:
- Incomplete or incorrect data can lead to misleading results.
- Symptoms can vary significantly among individuals leading to overlaps with multiple conditions.
- The effectiveness of the model relies heavily on the quality of generated embeddings.
- Different descriptions of symptoms by users can complicate matching with given symptom descriptions.
- Handling sensitive health-related data raises concerns about patient confidentiality and data security.
Conclusion
In this article, we used small language models to enhance healthcare accessibility and efficiency through a disease diagnosis system based on symptom analysis. By using embeddings from a small language model the system can identify conditions based on user input symptoms providing us with treatment recommendations. Addressing challenges related to data quality, symptom ambiguity and user input variability is essential for improving accuracy and user experience.
Key Takeaways
- Embedding models like MiniLM-L6-V2 enable precise symptom analysis and healthcare recommendations.
- Compact small language models efficiently support healthcare AI on resource-constrained devices.
- High-quality embedding generation is crucial for accurate symptom and condition matching.
- Addressing data quality and variability enhances reliability in AI-driven health recommendations.
- The system’s effectiveness hinges on robust data handling and diverse symptom descriptions.
Frequently Asked Questions
A. The system helps the user identify potential medical conditions based on reported symptoms by comparing them to a database of known conditions.
A. It uses a pre-trained Sentence Transformer model MiniLM-L6-V2 to convert symptoms into vector embeddings capturing their semantic meanings for better comparison.
A. While it provides useful insights it cannot replace professional medical advice and may struggle with vague symptom descriptions limited by the quality of its underlying dataset.
A. Accuracy varies based on input quality and underlying data. Results should be considered preliminary before consulting a healthcare professional.
A. Yes, it accepts a string of symptoms though matching effectiveness may depend on how clearly the symptoms are expressed.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.






