This article was published as a part of the Data Science Blogathon
Let’s learn about the pre-trained stacked model and detect if the person has Pneumonia or not.
Introduction
Computer Vision is taking over the world, tasks that were previously handled by humans themselves are now being done via computer in many fields. One of the areas where computer vision is being applied is Healthcare. Today we will see one of the major use cases in the healthcare industry i.e. Pneumonia detection in images. Pneumonia is an infectious lung disease. Bacteria, viruses, and fungi cause it. This causes inflammation in the air sacs in your lungs, which are called alveoli. when alveoli get filled with fluid or pus, it makes it difficult to breathe. Detecting Pneumonia from X-Ray pictures is a finely detailed image classification, in this case, we need to take each and every minor detail into training.
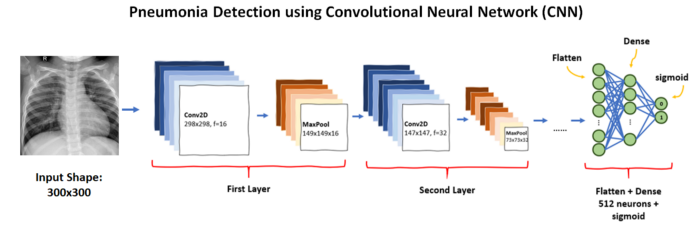
Training a CNN model gives somewhere around 65% accuracy hence we will use pre-trained stacked models.
About Dataset
- The dataset contains training data, validation data, and testing data.
- The training data contains 5,216 total x-ray images with 3,875 images to have pneumonia and 1,341 images to be normal.
- The validation data is comparatively smaller with only 16 images with 8 cases of pneumonia and 8 normal cases.
- The testing data consists of 624 images split between 390 pneumonia cases and 234 normal cases
- Dataset can be downloaded from this link.
Let’s get started!
Importing Libraries
As always let’s start with importing the libraries.
from keras.layers.merge import concatenate from keras.layers import Input import tensorflow as tf from tensorflow.keras.optimizers import Adam from tensorflow.keras.preprocessing.image import ImageDataGenerator from tensorflow.keras.layers import Conv2D, MaxPooling2D,GlobalAveragePooling2D, Flatten, Dense, Dropout, BatchNormalization from tensorflow.keras.applications.densenet import DenseNet169
from tensorflow.keras.applications.densenet import preprocess_input as densenet_preprocess
from tensorflow.keras.applications.mobilenet_v2 import MobileNetV2
Loading the Data
Our dataset have hierarchical folders for the training and validation so we have segregated the path for loading the data
- train_n:
- train_p:
- validation_data_dir: validation data
- test_data_dir: for testing
main_dir = "../input/chest-xray-pneumonia/chest_xray/" train_data_dir = main_dir + "train/" validation_data_dir = main_dir + "val/" test_data_dir = main_dir + "test/" train_n = train_data_dir+'NORMAL/' train_p = train_data_dir+'PNEUMONIA/'
Fitting Data Generators
We need a data generator because training a big amount of images can take us to memory insufficient error.
In order to avoid overfitting problems, we need to expand artificially our dataset, we can make our existing dataset even larger but the idea is to alter the training data with a small transformation to reproduce the variations.
Here data generator will do all our image processing tasks for training.
Note– Here we will apply image augmentation only on the training images, not on testing or validation.
train_datagen = ImageDataGenerator(
rescale=1. / 255,
shear_range=0.2,
zoom_range=0.2,
width_shift_range=0.1,
height_shift_range=0.1,
brightness_range=[0.2,1.0],
horizontal_flip=True)
test_datagen = ImageDataGenerator(rescale=1. / 255)
val_datagen = ImageDataGenerator(rescale=1. / 255)
We have applied necessary image preprocessing ie- rescaling, horizontal flip, brightness range, width_shift_range, shear_range, etc.
- brightness_range:
- shear_range:
- zoom_range:
- rescale:
Loading Data Generators
The flow_from_directory loads the data
into the data-generator by taking the data path. it loads the data by
going to all folders in a hierarchical manner.
img_width , img_height = [224,224]
batch_size = 16train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary',
shuffle = True)
validation_generator = val_datagen.flow_from_directory(
validation_data_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode='binary')
test_generator = test_datagen.flow_from_directory(
test_data_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode='binary')
class_mode = ‘binary’ since its a binary classification.
target_size = (img_height, img_width) target image size.
batch_size = 16 data will be generated in batch form, in order to avoid memory insufficiency errors.
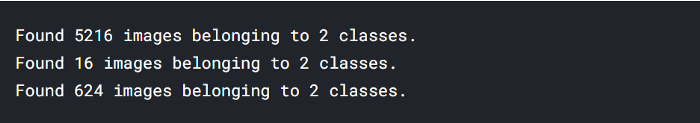
- 5216 images belonging to 2 classes (training data)
- 16 images belonging to 2 classes (validation data)
- 624 images belonging to 2 classes (testing data)
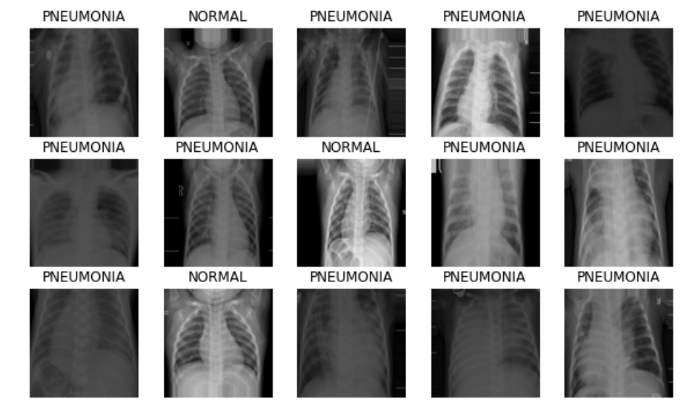
Further, we can plot the data batches generated by our data generator.
image_batch, label_batch = next(iter(train_generator))
def show_batch(image_batch, label_batch):
plt.figure(figsize=(10, 10))
for n in range(15):
ax = plt.subplot(5, 5, n + 1)
plt.imshow(image_batch[n])
if label_batch[n]:
plt.title("PNEUMONIA")
else:
plt.title("NORMAL")
plt.axis("off")
show_batch(image_batch, label_batch)
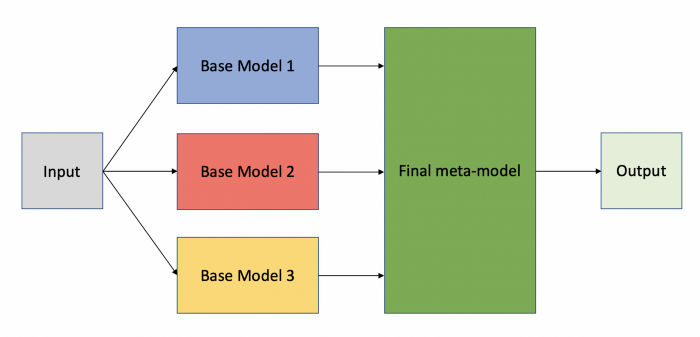
Pre-trained Stacked Model Designing
We will stack DenseNet169 and MobilenetV2 for improved results, stacking 2 models: freezing initial layers and then stacking the output.
input_shape = (224,224,3)
input_layer = Input(shape = (224, 224, 3))
#first model
base_mobilenet = MobileNetV2(weights = 'imagenet', include_top = False, input_shape = input_shape)
base_densenet = DenseNet169(weights = 'imagenet', include_top = False, input_shape = input_shape)for layer in base_mobilenet.layers:
layer.trainable = False
for layer in base_densenet.layers:
layer.trainable = False
model_mobilenet = base_mobilenet(input_layer)
model_mobilenet = GlobalAveragePooling2D()(model_mobilenet)
output_mobilenet = Flatten()(model_mobilenet)
model_densenet = base_densenet(input_layer)
model_densenet = GlobalAveragePooling2D()(model_densenet)
output_densenet = Flatten()(model_densenet)
merged = tf.keras.layers.Concatenate()([output_mobilenet, output_densenet])
x = BatchNormalization()(merged)
x = Dense(256,activation = 'relu')(x)
x = Dropout(0.5)(x)
x = BatchNormalization()(x)
x = Dense(128,activation = 'relu')(x)
x = Dropout(0.5)(x)
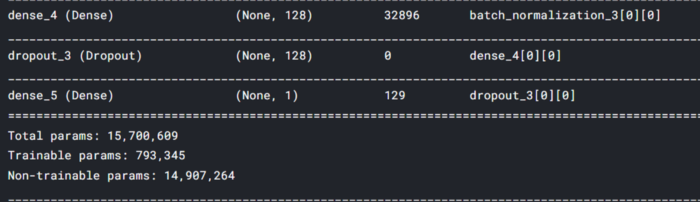
x = Dense(1, activation = 'sigmoid')(x)
stacked_model = tf.keras.models.Model(inputs = input_layer, outputs = x)
We are using functional API to design our model’s layers.
Freezing all the intermediate layers to keep the pre-trained information and only train on the last layers after stacking.
These pre-trained stacked models are trained on the imagenet dataset.
tf.keras.layers.Concatenate() concatenate the outputs of the models.
stacked_model is our final stacked model ready for training.
Model Compilation:
We are using adam optimizer with LR = 0.0001.
optm = Adam(lr=0.0001)
stacked_model.compile(loss='binary_crossentropy', optimizer=optm,
metrics=['accuracy'])
Defining Callbacks
Callbacks aren’t always mandatory but it always helps to train our model efficiently.
from keras.callbacks import EarlyStopping,ReduceLROnPlateau
EarlyStopping = EarlyStopping(monitor='val_accuracy',
min_delta=.01,
patience=6,
verbose=1,
mode='auto',
baseline=None,
restore_best_weights=True)
EarlyStopping: if the pre-trained stacked model doesn’t improve further training will stop after patience epochs.
rlr = ReduceLROnPlateau( monitor="val_accuracy",
factor=0.01,
patience=6,
verbose=0,
mode="max",
min_delta=0.01)
ReduceLROnPlateau reduces the learning rate if the model doesn’t improve.
model_save = ModelCheckpoint('./stacked_model.h5',
save_best_only = True,
save_weights_only = False,
monitor = 'val_loss',
mode = 'min', verbose = 1)
ModelCheckpoint: it saves the best-performed epochs.
Pre-trained Stacked Model Training
Training the pre-trained stacked model using our defined data generator.
nb_train_samples = 5216 # number of training samples nb_validation_samples = 16 # number of validation samples nb_test_samples = 624 # number of training samples epochs = 20 # number of epochs we gonna run batch_size = 16 # batch size ( at every iteration it will take 16 batches for training)
Calling the fit method calls the training on our model.
stacked_history = stacked_model.fit(train_generator,
steps_per_epoch = nb_train_samples // batch_size,
epochs = 20,
validation_data = test_generator,
callbacks=[EarlyStopping, model_save,rlr])
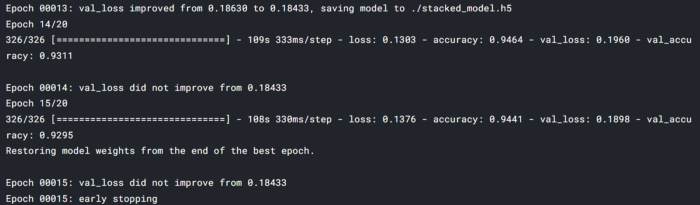
maximum val_accuracy is 93% and loss is .18 the minimum loss. we can further do some fine-tuning and improve the performance.
Pre-trained Stacked Model Testing
Defining function for prediction, plotting results, and testing.
def process_image(image):
image = image/255
image = cv2.resize(image, (224,224))
return image
preprocess_image() takes image array and rescales and resizes it to (224,224).
def predict(image_path, model):
im = cv2.imread(image_path)
test_image = np.asarray(im)
processed_test_image = process_image(test_image)
processed_test_image = np.expand_dims(processed_test_image, axis = 0)
ps = model.predict(processed_test_image)
return ps
predict() inputs image_path and model and returns prediction.
import seaborn as sns from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from PIL import Image
def testing(model, test_df):
base_pred =[]
for image in test_df.img_path:
base_pred.append(predict(image , model)[0][0])
final_base_pred = np.where(np.array(base_pred)>0.5,1,0)
actual_label = test_df['label']
# print(final_base_pred)
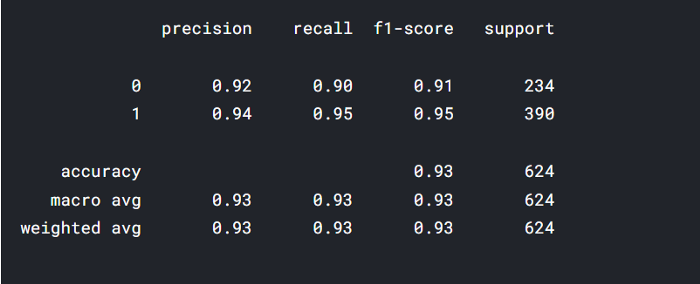
print("Classification Report:", classification_report(actual_label, final_base_pred))
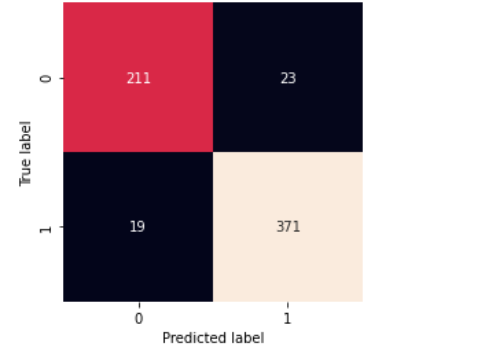
matrix=confusion_matrix(actual_label, final_base_pred)
sns.heatmap(matrix,square=True, annot=True, fmt='d', cbar=False,
xticklabels=['0', '1'],
yticklabels=['0', '1'])
plt.xlabel('Predicted label')
plt.ylabel('True label')
testing() takes dataset and model and returns confusion matrix along with classification_report.
testing(stacked_model,test_df)
Takeaway
- Our stacked model has the lowest loss in 20 epochs and the highest validation accuracy.
- Further, we can apply (CLAHE and normalization) for a better model.
- We can stack more models and can try to improve our model.
Conclusion
In this article, we saw how to design a pre-trained stacked model for pneumonia detection and got a good result. A simple deep learning model can also be used for such tasks but that totally depends on the data if you feel that your data is a bit complex you can go ahead with stacked models otherwise stacked models are preferred.
Stacking models give better results for finely detailed image classification. Want to read more about pre-trained stacked models? Click here.
Thanks for reading the article, please share below if you liked this article.
Reach out to me on LinkedIn.





