This article was published as a part of the Data Science Blogathon.
Introduction
Working on a task involving the interpretation of chest X-ray medical images and no labeled data at your disposal? Well, no problem. Researchers from Harvard Medical School and Stanford University have devised an artificial intelligence diagnostic tool that can detect diseases from natural language descriptions of chest X-rays without needing the labeled data.
This is a major step toward significant advancement in clinical AI design because most existing models require vast amounts of annotated data before that data can be fed into a model for training.
Source: Canva
This research paper will look at the proposed method in further detail. Let’s dive right in!
Highlights
-
Researchers from Harvard Medical School and Stanford University have devised an artificial intelligence diagnostic tool (ChexZero) that can detect diseases from natural language descriptions of chest X-rays without needing the labeled data.
-
Zero-shot learning was utilized to classify pathologies in chest X-rays rather than training the model on explicit labels. To do this, the model learned to predict which chest X-ray corresponds to which radiology report using image-text pairs of chest X-rays and radiology reports.
-
On an external validation dataset of chest X-rays, the CheXzero model outperformed a fully supervised model in the detection of three pathologies (out of eight), and the performance generalized to pathologies that were not explicitly annotated for model training, multiple image-interpretation tasks, and to datasets from multiple institutions.
Background of CheXzero
To achieve a high level of performance, the models are typically trained on relevant datasets that professionals have carefully annotated. Several approaches, including model pre-training and self-supervision, have been put forth to lessen the dependence of models on large labeled datasets.
Self-supervised pre-training techniques have proven to increase label efficiency across a range of medical tasks. However, they still require a supervised fine-tuning step after pre-training that requires manually labeled data for the model to predict relevant pathologies accurately. Hence, these approaches can only predict diseases explicitly annotated in the training dataset and can’t predict pathologies not explicitly annotated for training.
Given those above, for the model to predict a certain pathology with decent performance, it must be provided with a good number of expert-labeled training examples for that pathology during training. This process of obtaining high-quality annotations of certain pathologies can be expensive and time-consuming, frequently leading to major inefficiencies in clinical artificial intelligence workflows.
Methodology of CheXzero
To address the limitations above, a machine-learning paradigm is used where a model can classify samples during testing that were not explicitly annotated during training. To build a model with high performance for the multi-label classification of chest X-ray images, researchers presented a zero-shot method using a fully self-supervised learning technique. This method does not require explicit manual/annotated chest X-ray image interpretation labels.
The method, called CheXzero, uses contrastive learning, a kind of self-supervised learning, with image–text pairs to learn a representation that facilitates zero-shot multi-label classification.
In contrast to previous self-supervised approaches, this method can accurately identify pathologies that were not explicitly annotated and don’t require fine-tuning using labeled data except for testing.
Training pipeline: To devise the method, researchers utilized that radiology images are naturally labeled through corresponding clinical reports and that these reports can provide a natural source of supervision.
Zero-shot learning was utilized to classify pathologies in chest X-rays rather than training the model on explicit labels (Figure. 1). To do this, the model learned to predict which chest X-ray corresponds to which radiology report using image-text pairs of chest X-rays and radiology reports. Notably, the model was trained with 377,110 pairs of chest X-ray images and the corresponding radiology report (raw) from the MIMIC-CXR dataset.
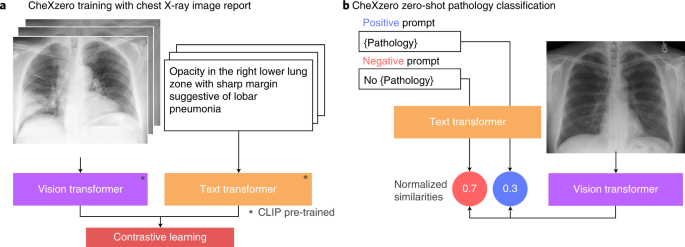
Figure1: a) CheXzero training with chest X-ray image report b) CheXzero zero-shot pathology classification.
Zero-shot Pathology Classification: A positive and negative prompt (like ‘consolidation’ versus ‘no consolidation’) was generated for each pathology. By assessing the model output for the positive and negative prompts, the self-supervised method estimates a probability score for the pathology, which can be used to classify its presence in the chest X-ray image.
On testing, it was discovered that CheXzero was capable of identifying pathologies that humans didn’t specifically annotate. Moreover, It performed better than other self-supervised AI tools and had accuracy comparable to that of human radiologists.
Ekin Tiu, who co-authored the study, claims that CheXzero has eliminated the need for fairly large labeled datasets to interpret complex medical images. CheXzero can generalize to various imaging modalities (CT scans, MRIs, and echocardiograms) in medical settings with unstructured data. It holds the promise of getting around the large-scale labeling bottleneck in medical machine learning.
Conclusion
To sum it up, in this article, we learned the following:
- Researchers from Harvard Medical School and Stanford University have devised an artificial intelligence diagnostic tool, CheXzero, that can detect diseases from natural language descriptions of chest X-rays without needing the labeled data.
- Zero-shot learning was utilized to classify pathologies in chest X-rays rather than training the model on explicit labels. To do this, the model learned to predict which chest X-ray corresponds to which radiology report using image-text pairs of chest X-rays and radiology reports.
- A positive and negative prompt (like ‘consolidation’ versus ‘no consolidation’) was generated for each pathology. By assessing the model output for the positive and negative prompts, the self-supervised method estimates a probability score for the pathology, which can be used to classify its presence in the chest X-ray image.
- The self-supervised method outperformed previous label-efficient approaches to chest X-ray pathology classification, indicating that explicit labels are not necessary to perform well on medical image interpretation tasks when corresponding reports are available for training.
- CheXzero can generalize to various imaging modalities (CT scans, MRIs, and echocardiograms) in medical settings with unstructured data. It holds the promise of getting around the large-scale labeling bottleneck in medical machine learning.
That concludes this article. Thanks for reading. If you have any questions or concerns, please post them in the comments section below. Happy learning!
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.





