Introduction
Yes, you heard that right, there is a way pf making your python code run 1000x faster by using a library I recently discovered, called Numba. It is a JIT (Just in Time) compiler, which takes your existing python code, figures out what types it contains, and generates a very fast machine code. Their tagline is:
” Numba makes Python code fast “
the name “Numba” comes from “NumPy” + “Mamba” which is a clever wordplay because Mamba snakes are known to crunch their prey quicker and generally faster than Pythons. The machine code generated by Numba is as fast as languages like C, C++, and Fortran without having to code in those languages. Numba works really well with Numpy arrays, which is one of the reasons why it is used more and more in scientific computing. Here is a Fun fact, Numba is also used by astronomers, along with AstroPy, for numerical algorithms, focusing on how to get very good performance on the CPU.
Parallel computing can also be done with Numba. If you have a high-performance computer with GPU, you can go a step further and run algorithms parallelly in GPU too, generating much higher performance. This can exponentially decrease the time required for you to run simulations, train models faster as well as efficiently use your hardware.
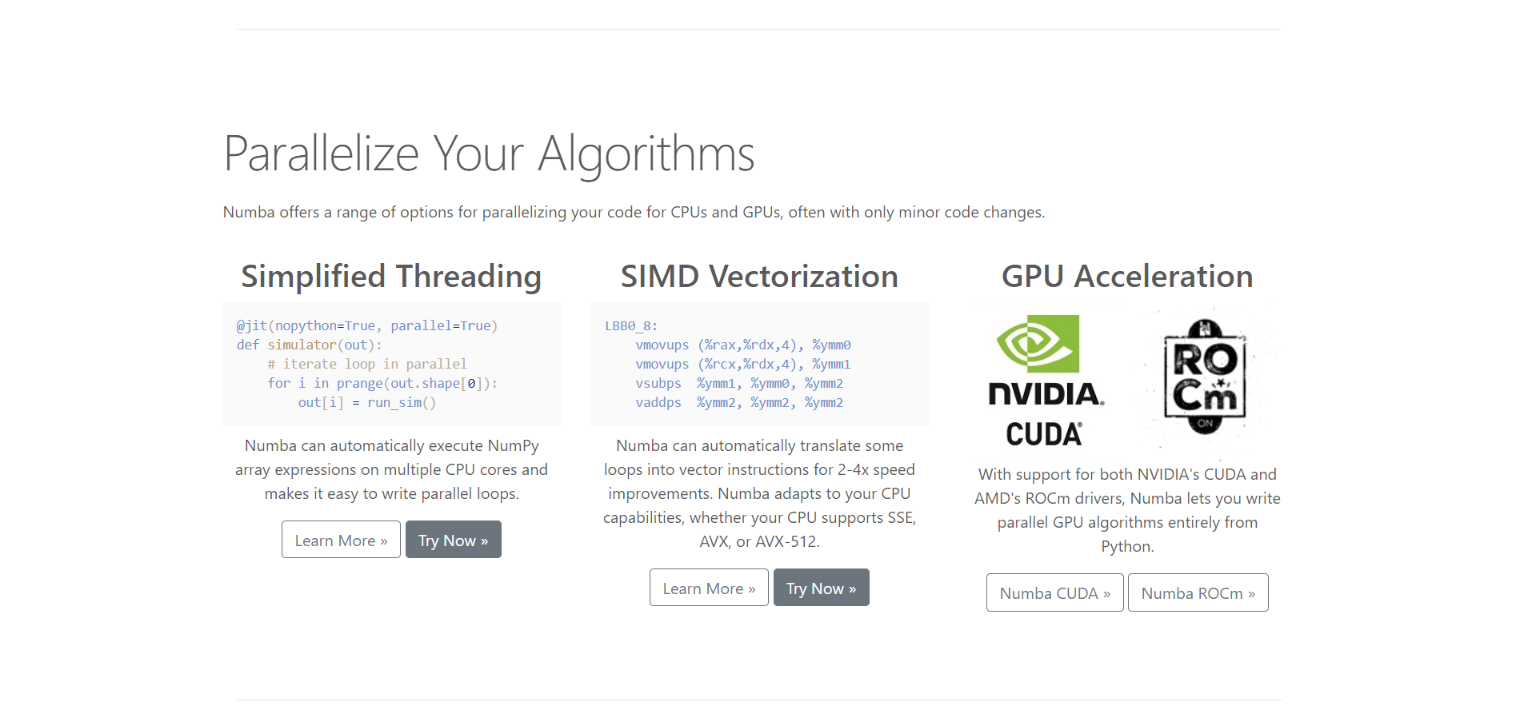
How Numba works
Let us dive into some examples to understand how Numba works. For the first example, we’ll use the code snippet provided on the official website of Numba. Here we’ll use the Monte Carlo algorithm to estimate the value of Pi. The algorithm basically works by generating and plotting a bunch of random numbers in a 2 by 2 square, and then check which ones are x2 + y2 < 1.0 i.e inside a 2r circle, inscribed in the square. Then calculate the ratio of the number of points inside the circle to the total number of points generated.
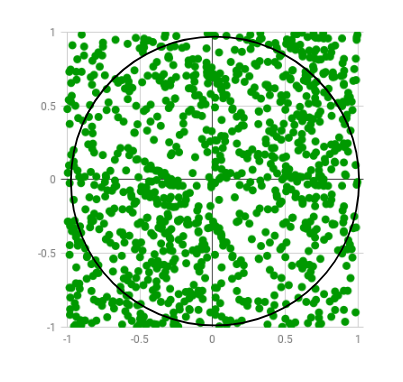
Image Source: GeeksForGeeks
First, let us import the necessary libraries
Here is the code for the Monte Carlo algorithm for estimating Pi
import random
from numba import jit,njit,vectorize
def monte_carlo_pi(nsamples):
acc = 0
for i in range(nsamples):
x = random.random()
y = random.random()
if (x ** 2 + y ** 2) < 1.0:
acc += 1
return 4.0 * acc / nsamplesHere we haven’t used the Numba decorator yet, let us see how the code performs:
%time monte_carlo_pi( 10000 )

So we can see that it takes the Python interpreter 7.41 ms to run the monte_carlo_pi function with a value of 10,000. Thus we can see that it takes a reasonable amount of time. Now let’s make a jitted version of the function, using Numba decorator jit(). We can create it by simply typing:
monte_carlo_pi_jit = jit() (monte_carlo_pi)
And now run the function
monte_carlo_pi_jit( 10000 )
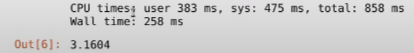
Huh, what do we have here? As you can see this is a surprising result. It took 258ms, which is almost 30 times what we had in the pure Python version. Well, this happens because Numba is a JIT compiler, so around the first time it’ll spend some extra moments compiling the code, and now since you’ve run the function once and all that code has been cached, if you rerun it, or use it somewhere else in the program, the computation time will drastically decrease. Let’s rerun the above line of code and see the results:
monte_carlo_pi_jit( 10000 )
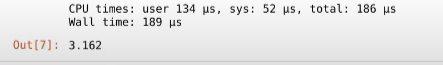
And voila! as you can see, the time it took is just 189 µs!! which is 0.189 ms, almost 1000 times as fast as our pure python code!
Limitations of Numba
Even roses have thorns. Turns out Numba has some small limitations and shortcomings which might throw you in a pickle now and then.
1. Numba compiles individual functions, not whole programs.
2. Numba supports a subset of Python. Some dict/list/set support but not mixed types of keys and values nor mixed types in lists.
3. Numba supports a subset of NumPy. Not all functions and methods are available.
4. Numba does not support Scikitlearn, PyData, and some other Python packages.
5. Works best with Numpy arrays than Python lists. But this is not much of a limitation, since most of our Data science computation is done with NumPy arrays.
More about Numba.Jit modes
Like I previously stated, Numba is mainly for numerical computing and does not really support string data types that well. But what happens if we use it with a string? Here is a simple function to test it out:
def original_function(input_list):
output_list = []
for item in input_list:
if item % 2 == 0:
output_list.append(2)
else:
output_list.append('1')
return output_list
test_list = list(range(10)
%time original_function(test_list)
> [2, '1', 2, '1', 2, '1', 2, '1', 2, '1']
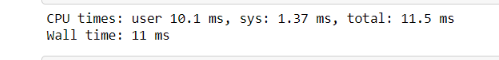
As you can see, we’ve appended “1” to the output_list which is a string type and a bug, and the list returned is a mixture of int and str types. Python has no problem handling this and does that taking a decent amount of time. Now let’s create a jitted version.
jitted_function = jit()(original_function)
%time jitted_function(test_list)
> NumbaWarning : Compilation is falling back to Object Mode.
What Numba essentially does is fall back to something called “Object Mode” which is due to the mismatch of data types, and this generates a warning. This mode ends up hurting you since it takes up much more time trying to be flexible and ends up taking more time than Pure python code.
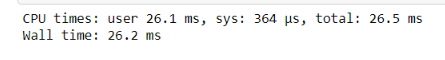
This has an easy fix, you can either use the @njit operator or the @jit(nopython = True). And now if we run this code, we’ll get an error, not a warning, which is good in the long run for a Data scientist developing a high-performance algorithm, because we meant to append 1, not “1” and its an obvious bug, and sometimes these bugs are not readily obvious so it’s better to get an error message.
@Vectorize decorator
What vectorized does is it allows us to rewrite this function as a scalar computation. What this essentially means is that we can rewrite our previous function, and this time we take just a single number(scalar) as input and apply the @vectorize wrapper. This new function can take both lists as well as a single-digit as input and produce the desired output.
@vectorize(nopython=True)
def non_list_function(item):
if item % 2 == 0:
return 2
else:
return 1
%time non_list_function(test_list)
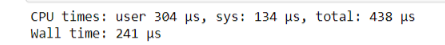
As you can see, this takes even less time than the @jit decorator.
Some more wrappers provided by Numba:
- @vectorize: allows scalar arguments to be used as NumPy
ufuncs, - @guvectorize: produces NumPy generalized
ufuncs, - @stencil: declare a function as a kernel for a stencil-like operation,
- @jitclass: for jit aware classes,
- @cfunc: declare a function for use as a native call back (to be called from C/C++ etc),
- @overload: register your own implementation of a function for use in nopython mode, e.g. @overload(scipy.special.j0).
Source: Numba
You can further read about this awesome library in their official documentation here.
Here are some of the video materials I found particularly useful:
1. Christopher Deil: Understanding NUmba, the Python and NumPy compiler
2. How to Accelerate an existing codebase with Numba | SciPy 2019 | Siu Kwan Lam, Stanley Seibert
The media shown in this article are not owned by Analytics Vidhya and is used at the Author’s discretion.





