This article was published as a part of the Data Science Blogathon
Anomaly Detection
Anomaly detection is the process of finding abnormalities in data. Abnormal data is defined as the ones that deviate significantly from the general behavior of the data. Some of the applications of anomaly detection include fraud detection, fault detection, and intrusion detection. Anomaly Detection is also referred to as outlier detection.
Some of the anomaly detection algorithms are,
- Local Outlier Factor
- Isolation Forest
- Connectivity Based Outlier Factor
- KNN based Outlier Detection
- One class SVM
- AutoEncoders
Outlier Detection vs Novelty Detection
In outlier detection, the training data consists of both anomalies and normal observations whereas in novelty detection the training data consists only of normal observations rather than having both normal and anomalous observations. In this post, we’re gonna see a use case of novelty detection.
AutoEncoder
AutoEncoder is an unsupervised Artificial Neural Network that attempts to encode the data by compressing it into the lower dimensions (bottleneck layer or code) and then decoding the data to reconstruct the original input. The bottleneck layer (or code) holds the compressed representation of the input data. The number of hidden units in the code is called code size.
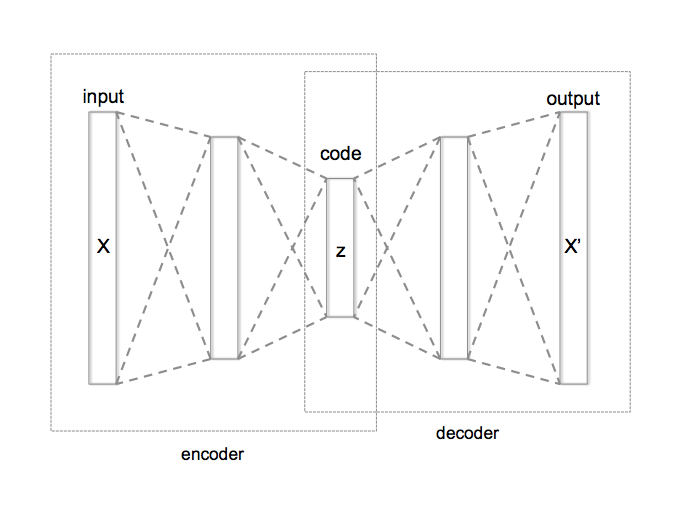
Applications of AutoEncoders
- Dimensionality reduction
- Anomaly detection
- Image denoising
- Image compression
- Image generation
In this post let us dive deep into anomaly detection using autoencoders.
Anomaly Detection using AutoEncoders
AutoEncoders are widely used in anomaly detection. The reconstruction errors are used as the anomaly scores. Let us look at how we can use AutoEncoder for anomaly detection using TensorFlow.
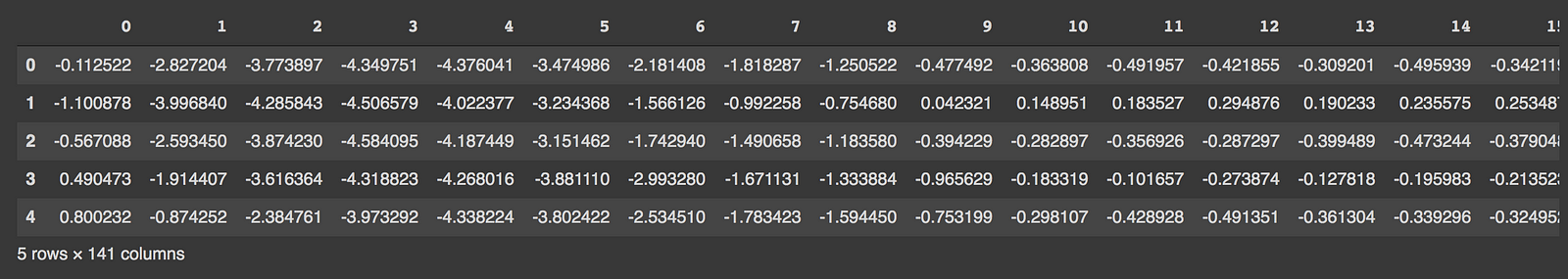
Import the required libraries and load the data. Here we are using the ECG data which consists of labels 0 and 1. Label 0 denotes the observation as an anomaly and label 1 denotes the observation as normal.
import numpy as np import pandas as pd import tensorflow as tf import matplotlib.pyplot as plt from sklearn.metrics import accuracy_score from tensorflow.keras.optimizers import Adam from sklearn.preprocessing import MinMaxScaler from tensorflow.keras import Model, Sequential from tensorflow.keras.layers import Dense, Dropout from sklearn.model_selection import train_test_split from tensorflow.keras.losses import MeanSquaredLogarithmicError # Download the dataset PATH_TO_DATA = 'http://storage.googleapis.com/download.tensorflow.org/data/ecg.csv' data = pd.read_csv(PATH_TO_DATA, header=None) data.head() # data shape # (4998, 141)
# last column is the target
# 0 = anomaly, 1 = normal
TARGET = 140
features = data.drop(TARGET, axis=1)
target = data[TARGET]
x_train, x_test, y_train, y_test = train_test_split(
features, target, test_size=0.2, stratify=target
)
# use case is novelty detection so use only the normal data
# for training
train_index = y_train[y_train == 1].index
train_data = x_train.loc[train_index]
# min max scale the input data
min_max_scaler = MinMaxScaler(feature_range=(0, 1))
x_train_scaled = min_max_scaler.fit_transform(train_data.copy())
x_test_scaled = min_max_scaler.transform(x_test.copy())
The last column in the data is the target ( column name is 140). Split the data for training and testing and scale the data using MinMaxScaler.
# create a model by subclassing Model class in tensorflow
class AutoEncoder(Model):
"""
Parameters
----------
output_units: int
Number of output units
code_size: int
Number of units in bottle neck
"""
def __init__(self, output_units, code_size=8):
super().__init__()
self.encoder = Sequential([
Dense(64, activation='relu'),
Dropout(0.1),
Dense(32, activation='relu'),
Dropout(0.1),
Dense(16, activation='relu'),
Dropout(0.1),
Dense(code_size, activation='relu')
])
self.decoder = Sequential([
Dense(16, activation='relu'),
Dropout(0.1),
Dense(32, activation='relu'),
Dropout(0.1),
Dense(64, activation='relu'),
Dropout(0.1),
Dense(output_units, activation='sigmoid')
])
def call(self, inputs):
encoded = self.encoder(inputs)
decoded = self.decoder(encoded)
return decoded
model = AutoEncoder(output_units=x_train_scaled.shape[1])
# configurations of model
model.compile(loss='msle', metrics=['mse'], optimizer='adam')
history = model.fit(
x_train_scaled,
x_train_scaled,
epochs=20,
batch_size=512,
validation_data=(x_test_scaled, x_test_scaled)
)
The encoder of the model consists of four layers that encode the data into lower dimensions. The decoder of the model consists of four layers that reconstruct the input data.
The model is compiled with Mean Squared Logarithmic loss and Adam optimizer. The model is then trained with 20 epochs with a batch size of 512.
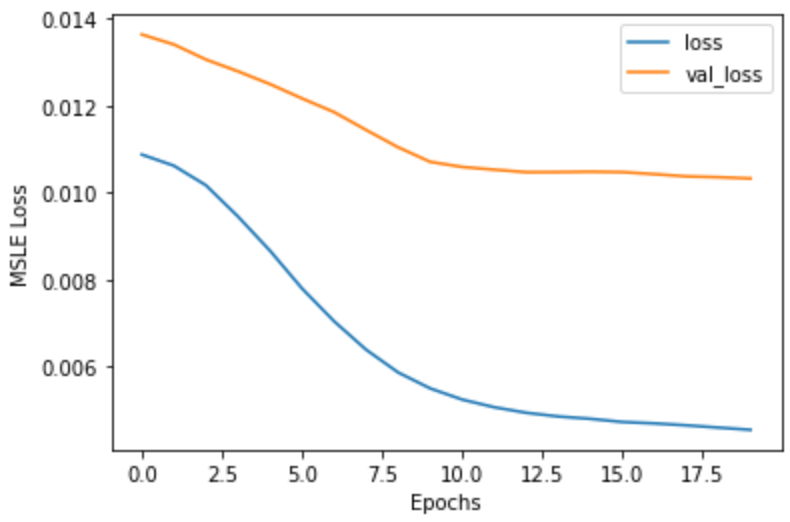
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.xlabel('Epochs')
plt.ylabel('MSLE Loss')
plt.legend(['loss', 'val_loss'])
plt.show()
def find_threshold(model, x_train_scaled):
reconstructions = model.predict(x_train_scaled)
# provides losses of individual instances
reconstruction_errors = tf.keras.losses.msle(reconstructions, x_train_scaled)
# threshold for anomaly scores
threshold = np.mean(reconstruction_errors.numpy()) \
+ np.std(reconstruction_errors.numpy())
return threshold
def get_predictions(model, x_test_scaled, threshold):
predictions = model.predict(x_test_scaled)
# provides losses of individual instances
errors = tf.keras.losses.msle(predictions, x_test_scaled)
# 0 = anomaly, 1 = normal
anomaly_mask = pd.Series(errors) > threshold
preds = anomaly_mask.map(lambda x: 0.0 if x == True else 1.0)
return preds
threshold = find_threshold(model, x_train_scaled)
print(f"Threshold: {threshold}")
# Threshold: 0.01001314025746261
predictions = get_predictions(model, x_test_scaled, threshold)
accuracy_score(predictions, y_test)
# 0.944
The reconstruction errors are considered to be anomaly scores. The threshold is then calculated by summing the mean and standard deviation of the reconstruction errors. The reconstruction errors above this threshold are considered to be anomalies. We can further fine-tune the model by leveraging Keras-tuner.
The autoencoder model does not have to symmetric encoder and decoder but the code size has to be smaller than that of the features in the data.
Find the entire code in my Google Colab Notebook.
References
Happy Deep Learning!
Thank You!
The media shown in this article are not owned by Analytics Vidhya and is used at the Author’s discretion.

