This article was published as a part of the Data Science Blogathon
SLR in Python with statsmodels.api, statsmodels.formula.api, and scikit-learn
In this blog, we will
- learn the basics of the Regression algorithm.
- take a sample dataset, perform EDA(Exploratory Data Analysis) and implement SLR(Simple Linear Regression) using statsmodels.api, statsmodels.formula.api, and scikit-learn.
To begin with, what is Regression Algorithm?
Regression is a ‘Supervised machine learning’ algorithm used to predict continuous features.
Linear regression is the simplest regression algorithm that attempts to model the relationship between dependent variable and one or more independent variables by fitting a linear equation/best fit line to observed data.
Based on the number of input features, Linear regression could be of two types:
- Simple Linear Regression (SLR)
- Multiple Linear Regression (MLR)
In Simple Linear Regression (SLR), we will have a single input variable based on which we predict the output variable. Where in Multiple Linear Regression (MLR), we predict the output based on multiple inputs.
Input variables can also be termed as Independent/predictor variables, and the output variable is called the dependent variable.
The equation for SLR is y=βo,+β1x+ϵ, where, Y is the dependent variable, X is the predictor, βo, β1 are coefficients/parameters of the model, and Epsilon(ϵ) is a random variable called Error Term.
OLS(Ordinary Least Squares), Gradient Descent are the two common algorithms to find the right coefficients for the minimum sum of squared errors.
Let’s begin by taking a small problem statement.
Problem statement: Build a simple linear regression model to predict the Salary Hike using Years of Experience.
Start by Importing necessary libraries
necessary libraries are pandas, NumPy to work with data frames, matplotlib, seaborn for visualizations, and sklearn, statsmodels to build regression models.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
from scipy import stats
from scipy.stats import probplot
import statsmodels.api as sm
import statsmodels.formula.api as smf
from sklearn import preprocessing
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_scoreOnce, we are done with importing libraries, we create a pandas dataframe from CSV file
df = pd.read_csv(“Salary_Data.csv”)
Perform EDA (Exploratory Data Analysis)
The basic steps of EDA are:
- Understand the dataset
- Identifying the number of features or columns
- Identifying the features or columns
- Identify the size of the dataset
- Identifying the data types of features
- Checking if the dataset has empty cells
- Identifying the number of empty cells by features or columns
- Handling Missing Values and Outliers
- Encoding Categorical variables
- Graphical Univariate Analysis, Bivariate
- Normalization and Scaling
Our dataset has two columns: YearsExperience, Salary. And both are of float datatype. We have 30 records and no null-values or outliers in our dataset.
Graphical Univariate analysis
For univariate analysis, we have Histogram, density plot, boxplot or violinplot, and Normal Q-Q plot. They help us understand the distribution of the data points and the presence of outliers.
A violin plot is a method of plotting numeric data. It is similar to a box plot, with the addition of a rotated kernel density plot on each side.
Python Code:
# Histogram
# We can use either plt.hist or sns.histplot
plt.figure(figsize=(20,10))
plt.subplot(2,4,1)
plt.hist(df['YearsExperience'], density=False)
plt.title("Histogram of 'YearsExperience'")
plt.subplot(2,4,5)
plt.hist(df['Salary'], density=False)
plt.title("Histogram of 'Salary'")
# Density plot
plt.subplot(2,4,2)
sns.distplot(df['YearsExperience'], kde=True)
plt.title("Density distribution of 'YearsExperience'")
plt.subplot(2,4,6)
sns.distplot(df['Salary'], kde=True)
plt.title("Density distribution of 'Salary'")
# boxplot or violin plot
# A violin plot is a method of plotting numeric data. It is similar to a box plot,
# with the addition of a rotated kernel density plot on each side
plt.subplot(2,4,3)
# plt.boxplot(df['YearsExperience'])
sns.violinplot(df['YearsExperience'])
# plt.title("Boxlpot of 'YearsExperience'")
plt.title("Violin plot of 'YearsExperience'")
plt.subplot(2,4,7)
# plt.boxplot(df['Salary'])
sns.violinplot(df['Salary'])
# plt.title("Boxlpot of 'Salary'")
plt.title("Violin plot of 'Salary'")
# Normal Q-Q plot
plt.subplot(2,4,4)
probplot(df['YearsExperience'], plot=plt)
plt.title("Q-Q plot of 'YearsExperience'")
plt.subplot(2,4,8)
probplot(df['Salary'], plot=plt)
plt.title("Q-Q plot of 'Salary'")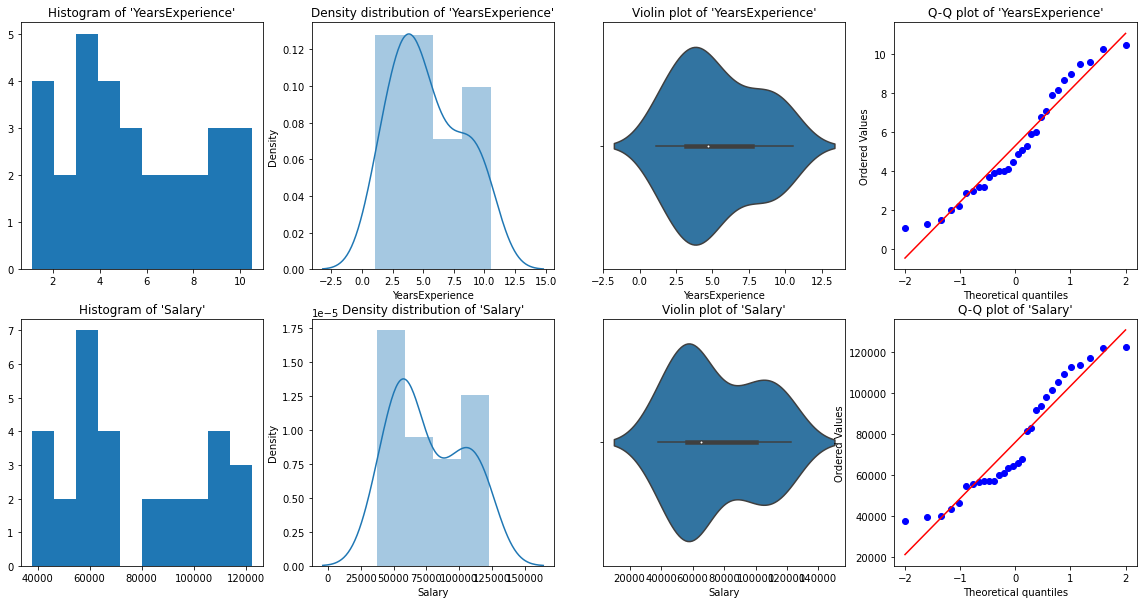
From the above graphical representations, we can say there are no outliers in our data, and YearsExperience looks like normally distributed, and Salary doesn't look normal. We can verify this using Shapiro Test.
Python Code:
# Def a function to run Shapiro test
# Defining our Null, Alternate Hypothesis
Ho = 'Data is Normal'
Ha = 'Data is not Normal'
# Defining a significance value
alpha = 0.05
def normality_check(df):
for columnName, columnData in df.iteritems():
print("Shapiro test for {columnName}".format(columnName=columnName))
res = stats.shapiro(columnData)
# print(res)
pValue = round(res[1], 2)
# Writing condition
if pValue > alpha:
print("pvalue = {pValue} > {alpha}. We fail to reject Null Hypothesis. {Ho}".format(pValue=pValue, alpha=alpha, Ho=Ho))
else:
print("pvalue = {pValue} <= {alpha}. We reject Null Hypothesis. {Ha}".format(pValue=pValue, alpha=alpha, Ha=Ha))
# Drive code
normality_check(df)Our instinct from the graphs was correct. YearsExperience is normally distributed, and Salary isn’t normally distributed.
Bivariate visualization
for Numerical vs. Numerical data, we can plot the below graphs
- Scatterplot
- Line plot
- Heatmap for correlation
- Joint plot
Python Code for various plots:
# Scatterplot & Line plots
plt.figure(figsize=(20,10))
plt.subplot(1,3,1)
sns.scatterplot(data=df, x="YearsExperience", y="Salary", hue="YearsExperience", alpha=0.6)
plt.title("Scatter plot")
plt.subplot(1,3,2)
sns.lineplot(data=df, x="YearsExperience", y="Salary")
plt.title("Line plot of YearsExperience, Salary")
plt.subplot(1,3,3)
sns.lineplot(data=df)
plt.title('Line Plot')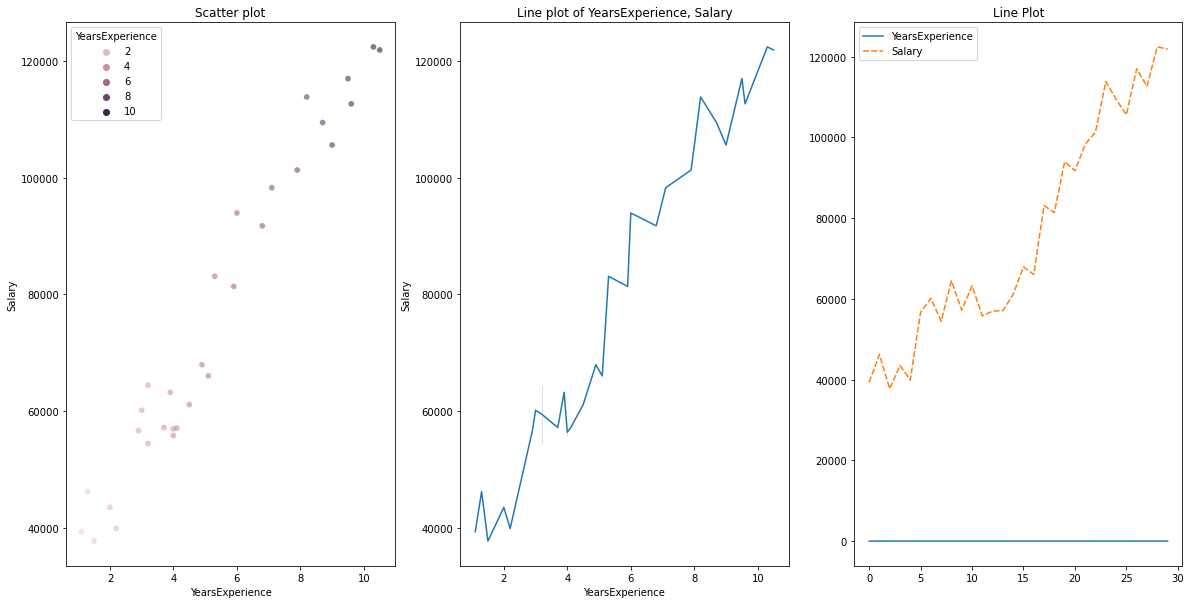
# heatmap
plt.figure(figsize=(10, 10))
plt.subplot(1, 2, 1)
sns.heatmap(data=df, cmap="YlGnBu", annot = True)
plt.title("Heatmap using seaborn")
plt.subplot(1, 2, 2)
plt.imshow(df, cmap ="YlGnBu")
plt.title("Heatmap using matplotlib")
# Joint plot
sns.jointplot(x = "YearsExperience", y = "Salary", kind = "reg", data = df)
plt.title("Joint plot using sns")
# kind can be hex, kde, scatter, reg, hist. When kind='reg' it shows the best fit line.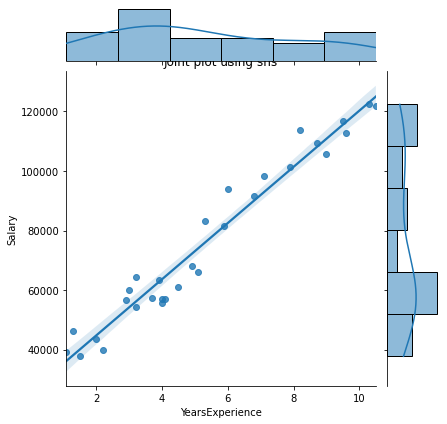
Check if there is any correlation between the variables using df.corr()
print("Correlation: "+ 'n', df.corr()) # 0.978 which is high positive correlation
# Draw a heatmap for correlation matrix
plt.subplot(1,1,1)
sns.heatmap(df.corr(), annot=True)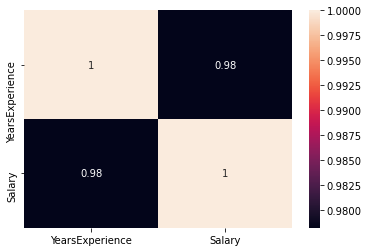
correlation =0.98, which is a high positive correlation. This means the dependent variable increases as the independent variable increases.
Normalization
As we can see, there is a huge difference between the values of YearsExperience, Salary columns. We can use Normalization to change the values of numeric columns in the dataset to use a common scale, without distorting differences in the ranges of values or losing information.
We use sklearn.preprocessing.Normalize to normalize our data. It returns values between 0 and 1.
# Create new columns for the normalized values
df['Norm_YearsExp'] = preprocessing.normalize(df[['YearsExperience']], axis=0)
df['Norm_Salary'] = preprocessing.normalize(df[['Salary']], axis=0)
df.head()Linear Regression using scikit-learn
LinearRegression(): LinearRegression fits a linear model with coefficients β = (β1, …, βp) to minimize the residual sum of squares between the observed targets in the dataset, and the targets predicted by the linear approximation.
def regression(df):
# defining the independent and dependent features
x = df.iloc[:, 1:2]
y = df.iloc[:, 0:1]
# print(x,y)
# Instantiating the LinearRegression object
regressor = LinearRegression()
# Training the model
regressor.fit(x,y)
# Checking the coefficients for the prediction of each of the predictor
print('n'+"Coeff of the predictor: ",regressor.coef_)
# Checking the intercept
print("Intercept: ",regressor.intercept_)
# Predicting the output
y_pred = regressor.predict(x)
# print(y_pred)
# Checking the MSE
print("Mean squared error(MSE): %.2f" % mean_squared_error(y, y_pred))
# Checking the R2 value
print("Coefficient of determination: %.3f" % r2_score(y, y_pred)) # Evaluates the performance of the model # says much percentage of data points are falling on the best fit line
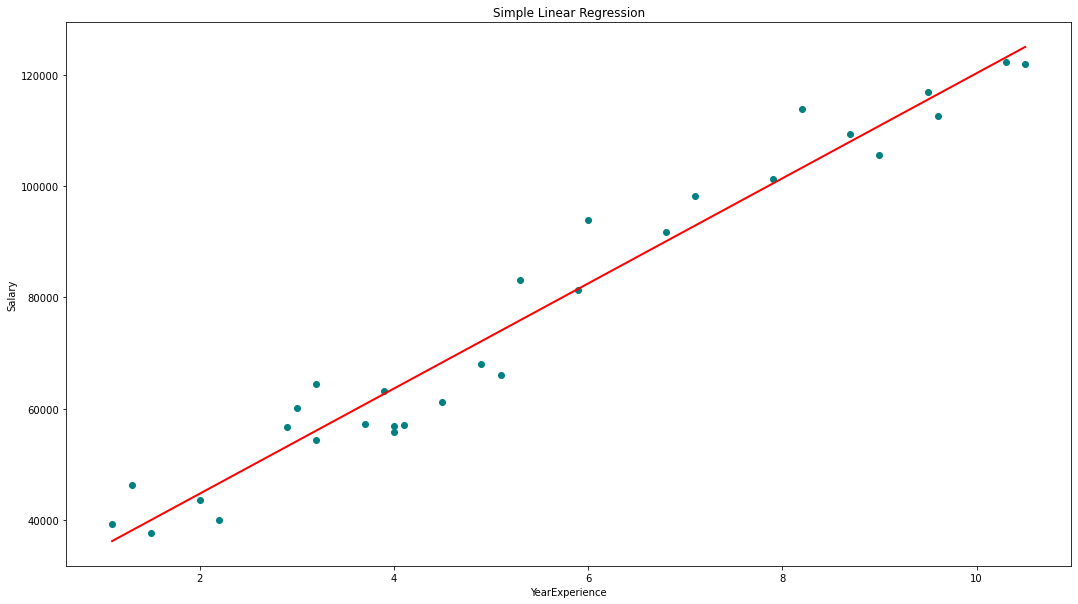
# visualizing the results.
plt.figure(figsize=(18, 10))
# Scatter plot of input and output values
plt.scatter(x, y, color='teal')
# plot of the input and predicted output values
plt.plot(x, regressor.predict(x), color='Red', linewidth=2 )
plt.title('Simple Linear Regression')
plt.xlabel('YearExperience')
plt.ylabel('Salary')
# Driver code
regression(df[['Salary', 'YearsExperience']]) # 0.957 accuracy
regression(df[['Norm_Salary', 'Norm_YearsExp']]) # 0.957 accuracyWe achieved 95.7% accuracy using scikit-learn but there is not much scope to understand the in-depth insights about the relevance of features from this model. So let’s build a model using statsmodels.api, statsmodels.formula.api
Linear Regression using statsmodel.formula.api (smf)
The predictors in the statsmodels.formula.api must be enumerated individually. And in this method, a constant is automatically added to the data.
def smf_ols(df):
# defining the independent and dependent features
x = df.iloc[:, 1:2]
y = df.iloc[:, 0:1]
# print(x)
# train the model
model = smf.ols('y~x', data=df).fit()
# print model summary
print(model.summary())
# Predict y
y_pred = model.predict(x)
# print(type(y), type(y_pred))
# print(y, y_pred)
y_lst = y.Salary.values.tolist()
# y_lst = y.iloc[:, -1:].values.tolist()
y_pred_lst = y_pred.tolist()
# print(y_lst)
data = [y_lst, y_pred_lst]
# print(data)
res = pd.DataFrame({'Actuals':data[0], 'Predicted':data[1]})
# print(res)
plt.scatter(x=res['Actuals'], y=res['Predicted'])
plt.ylabel('Predicted')
plt.xlabel('Actuals')
res.plot(kind='bar',figsize=(10,6))
# Driver code
smf_ols(df[['Salary', 'YearsExperience']]) # 0.957 accuracy
# smf_ols(df[['Norm_Salary', 'Norm_YearsExp']]) # 0.957 accuracy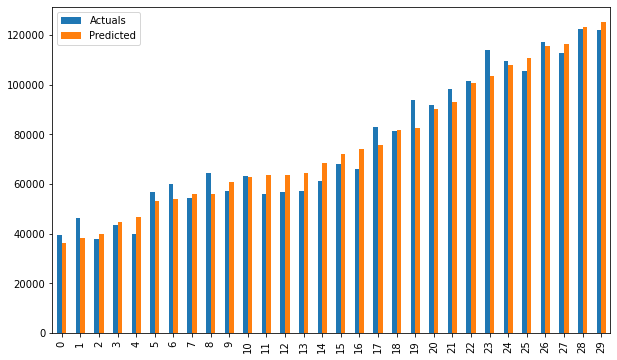
Bar plot of Actuals vs predicted values
Regression using statsmodels.api
The predictors are no longer have to be enumerated individually.
statsmodels.regression.linear_model.OLS(endog, exog)
endogis the dependent variableexogis the independent variable. An intercept is not included by default and should be added by the user(using add_constant).
# Create a helper function
def OLS_model(df):
# defining the independent and dependent features
x = df.iloc[:, 1:2]
y = df.iloc[:, 0:1]
# Add a constant term to the predictor
x = sm.add_constant(x)
# print(x)
model = sm.OLS(y, x)
# Train the model
results = model.fit()
# print('n'+"Confidence interval:"+'n', results.conf_int(alpha=0.05, cols=None)) #Returns the confidence interval of the fitted parameters. The default alpha=0.05 returns a 95% confidence interval.
print('n'"Model parameters:"+'n',results.params)
# print the overall summary of the model result
print(results.summary())
# Driver code
OLS_model(df[['Salary', 'YearsExperience']]) # 0.957 accuracy
OLS_model(df[['Norm_Salary', 'Norm_YearsExp']]) # 0.957 accuracyWe achieved 95.7% accuracy which is pretty good 🙂
What does the model summary table say??? 😕
It’s always important to understand certain terms from the regression model summary table so that we get to know the performance of our model and the relevance of the input variables.
Some important parameters that should be considered are the R-squared value, Adj. R-squared value, F-statistic, prob(F-statistic), coef of intercept and input variables, p>|t|.
- R-Squared is the coefficient of determination. A statistical measure that says much percentage of data points are falling on the best fit line. An R-squared value closer to 1 is expected for a model to fit well.
- Adj. R-squared penalizes the R-squared value if we keep adding the new features which are not contributing to the model prediction. If Adj. R-squared value < R-squared value, it’s a sign that we have irrelevant predictors in the model.
- F-statistic or F-test helps us to accept or reject Null Hypothesis. It compares the intercept-only model with our model with features. The null hypothesis is ‘all of the regression coefficients are equal to zero and that means both the models are equal’. The alternate hypothesis is ‘intercept the only model is worse than our model, which means our added coefficients improved the model performance. If prob(F-statistic) < 0.05 and F-statistic being a high value, we reject the Null hypothesis. It signifies that there is a good relationship between the input and the output variables.
- coef shows the estimated coefficients of the corresponding input features
- T-test talks about the relation between the output and each of the input variables individually. The null hypothesis is ‘coef of an input feature is 0’. The alternate hypothesis is ‘coef of an input feature is not 0’. If pvalue < 0.05, we reject the null hypothesis which indicates that there is a good relationship between the input variable and the output variable. We can eliminate the variables whose pvalue is >0.05.
Well, now we know how to draw important inferences from the model summary table, so now let’s look at our model parameters and evaluate our model.
In our case, the R-squared value (0.957) is close to Adj. R-squared value (0.955) is a good sign that the input features are contributing to the predictor model.
F-statistic is a high number and p(F-statistic) is almost 0, which means our model is better than the only intercept model.
pvalue of t-test for input variable is less than 0.05, so there is a good relationship between the input and the output variable.
Hence, we conclude by saying our model is performing well ✔😊
In this blog, we learned the basics of Simple Linear Regression (SLR), building a linear model using different python libraries, and drawing inferences from the summary table of OLS statsmodels.





