Photo by MART PRODUCTION from Pexels
Problem Statement:
To predict and localize brain tumors through image segmentation from the MRI dataset available in Kaggle.
I’ve divided this article into a series of two parts as we are going to train two deep learning models for the same dataset but the different tasks.
The model in this part is a classification model that will detect tumors from the MRI image and then if a tumor exists, we will further localize the segment of the brain having a tumor in the next part of this series.
Prerequisite:
Deep Learning
I’ll try to explicate every part thoroughly but in case you find any difficulty, let me know in the comment section. Let’s head into the implementation part using python.
Dataset: https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation
- First things first! Let us start by importing the required libraries.
import pandas as pd import numpy as np import seaborn as sns import matplotlib.pyplot as plt import cv2 from skimage import io import tensorflow as tf from tensorflow.python.keras import Sequential from tensorflow.keras import layers, optimizers from tensorflow.keras.applications.resnet50 import ResNet50 from tensorflow.keras.layers import * from tensorflow.keras.models import Model from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint from tensorflow.keras import backend as K from sklearn.preprocessing import StandardScaler %matplotlib inline
- Convert the CSV file of the dataset into a data frame to perform specific operations on it.
# data containing path to Brain MRI and their corresponding mask
brain_df = pd.read_csv('/Healthcare AI Datasets/Brain_MRI/data_mask.csv')
- View the DataFrame details.
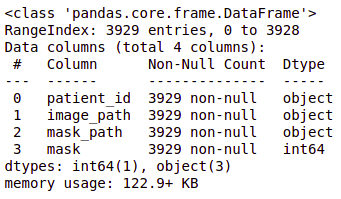
brain_df.info()
brain_df.head(5)
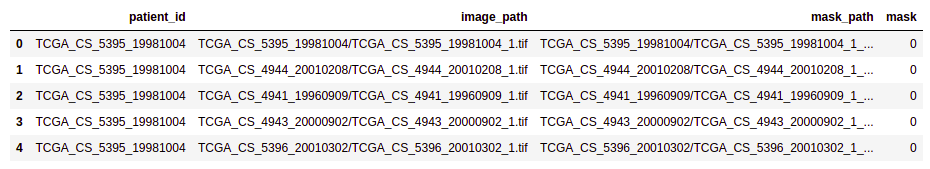
- Patient id: Patient id for each record (dtype: Object)
- Image path: Path to the image of the MRI (dtype: Object)
- Mask path: Path to the mask of the corresponding image (dtype: Object)
- Mask: Has two values: 0 and 1 depending on the image of the mask. (dtype: int64)
- Count the values of each class.
brain_df['mask'].value_counts()

- Randomly displaying an MRI image from the dataset.
image = cv2.imread(brain_df.image_path[1301]) plt.imshow(image)
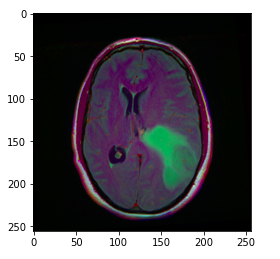
The image_path stores the path of the brain MRI so we can display the image using matplotlib.
Hint: The greenish portion in the above image can be considered as the tumor.
- Also, display the corresponding mask image.
image1 = cv2.imread(brain_df.mask_path[1301]) plt.imshow(image1)
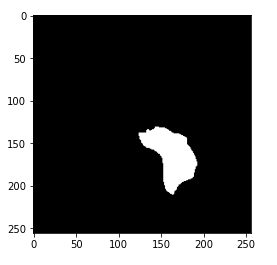
Now, you may have got the hint of what actually the mask is. The mask is the image of the part of the brain that is affected by a tumor of the corresponding MRI image. Here, the mask is of the above-displayed brain MRI.
- Analyze the pixel values of the mask image.
cv2.imread(brain_df.mask_path[1301]).max()
Output: 255
The maximum pixel value in the mask image is 255 which indicates the white color.
cv2.imread(brain_df.mask_path[1301]).min()
Output: 0
The minimum pixel value in the mask image is 0 which indicates the black color.
- Visualizing the Brain MRI, corresponding Mask, and MRI with the mask.
count = 0
fig, axs = plt.subplots(12, 3, figsize = (20, 50))
for i in range(len(brain_df)):
if brain_df['mask'][i] ==1 and count <5:
img = io.imread(brain_df.image_path[i])
axs[count][0].title.set_text('Brain MRI')
axs[count][0].imshow(img)
mask = io.imread(brain_df.mask_path[i])
axs[count][1].title.set_text('Mask')
axs[count][1].imshow(mask, cmap = 'gray')
img[mask == 255] = (255, 0, 0) #Red color
axs[count][2].title.set_text('MRI with Mask')
axs[count][2].imshow(img)
count+=1
fig.tight_layout()
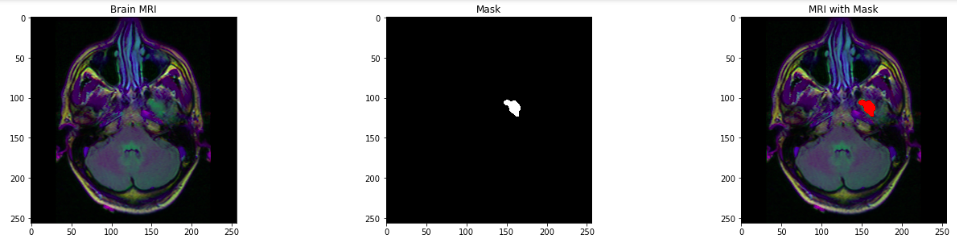
- Drop the id as it is not further required for processing.
# Drop the patient id column brain_df_train = brain_df.drop(columns = ['patient_id']) brain_df_train.shape
You will get the size of the data frame in the output: (3929, 3)
- Convert the data in the mask column from integer to string format as we will require the data in string format.
brain_df_train['mask'] = brain_df_train['mask'].apply(lambda x: str(x)) brain_df_train.info()
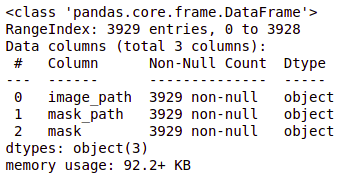
As you can see, now each feature has the datatype as an object.
- Split the data into train and test sets.
# split the data into train and test data from sklearn.model_selection import train_test_split train, test = train_test_split(brain_df_train, test_size = 0.15) |
- Augment more data using ImageDataGenerator. ImageDataGenerator generates batches of tensor image data with real-time data augmentation.
Refer here for more information on ImageDataGenerator and the parameters in detail.
We will create a train_generator and validation_generator from train data and a test_generator from test data.
# create an image generator from keras_preprocessing.image import ImageDataGenerator #Create a data generator which scales the data from 0 to 1 and makes validation split of 0.15 datagen = ImageDataGenerator(rescale=1./255., validation_split = 0.15) train_generator=datagen.flow_from_dataframe( dataframe=train, directory= './', x_col='image_path', y_col='mask', subset="training", batch_size=16, shuffle=True, class_mode="categorical", target_size=(256,256)) valid_generator=datagen.flow_from_dataframe( dataframe=train, directory= './', x_col='image_path', y_col='mask', subset="validation", batch_size=16, shuffle=True, class_mode="categorical", target_size=(256,256)) # Create a data generator for test images test_datagen=ImageDataGenerator(rescale=1./255.) test_generator=test_datagen.flow_from_dataframe( dataframe=test, directory= './', x_col='image_path', y_col='mask', batch_size=16, shuffle=False, class_mode='categorical', target_size=(256,256))
Now, we will learn the concept of Transfer Learning and ResNet50 Model which will be used for further training model.
Transfer Learning as the name suggests, is a technique to use the pre-trained models in your training. You can build your model on top of this pre-trained model. This is a process that helps you decrease the development time and increase performance.
ResNet (Residual Network) is the ANN trained on the ImageNet dataset that can be used to train the model on top of it. ResNet50 is the variant of the ResNet model which has 48 Convolution layers along with 1 MaxPool and 1 Average Pool layer.
- Here, we are using the ResNet50 Model which is a Transfer Learning Model. Using this, we will further add more layers to build our model.
# Get the ResNet50 base model (Transfer Learning) basemodel = ResNet50(weights = 'imagenet', include_top = False, input_tensor = Input(shape=(256, 256, 3))) basemodel.summary()
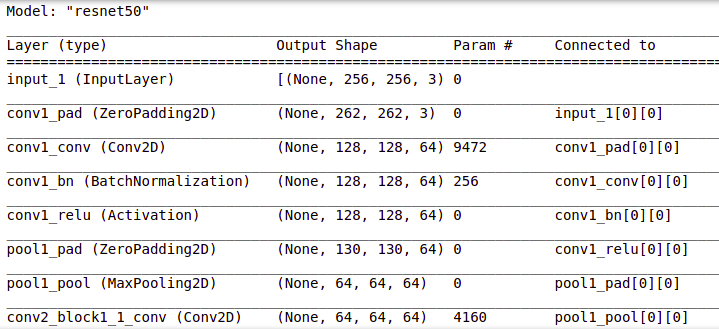
You can view the layers in the resnet50 model by using .summary() as shown above.
- Freeze the model weights. It means we will keep the weights constant so that it does not update further. This will avoid destroying any information during further training.
# freeze the model weights for layer in basemodel.layers: layers.trainable = False
- Now, as stated above we will add more layers on the top of the layers of ResNet50. These layers will learn to turn the old features into predictions on our dataset.
headmodel = basemodel.output headmodel = AveragePooling2D(pool_size = (4,4))(headmodel) headmodel = Flatten(name= 'flatten')(headmodel) headmodel = Dense(256, activation = "relu")(headmodel) headmodel = Dropout(0.3)(headmodel) headmodel = Dense(256, activation = "relu")(headmodel) headmodel = Dropout(0.3)(headmodel) headmodel = Dense(256, activation = "relu")(headmodel) headmodel = Dropout(0.3)(headmodel) headmodel = Dense(2, activation = 'softmax')(headmodel) model = Model(inputs = basemodel.input, outputs = headmodel) model.summary()
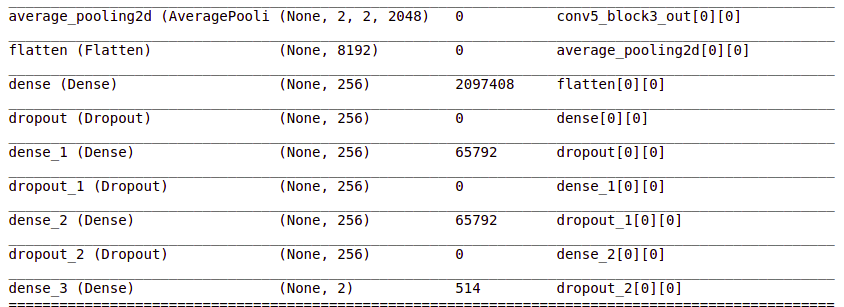
These layers are added and you can see them in the summary.
- Pooling layers are used to reduce the dimensions of the feature maps. The Average Pooling layer returns the average of the values.
- Flatten layers convert our data into a vector.
- A dense layer is the regular deeply connected neural network layer. Basically, it takes input and calculates output = activation(dot(input, kernel) + bias)
- The dropout layer prevents the model from overfitting. It randomly sets the input units of hidden layers to 0 during training.
- Compile the above-built model. Compile defines the loss function, the optimizer, and the metrics.
# compile the model model.compile(loss = 'categorical_crossentropy', optimizer='adam', metrics= ["accuracy"])
- Performing early stopping to save the best model with the least validation loss. Early stopping performs a large number of training epochs and stops training once the model performance does not further improve on the validation dataset.
Read more about EarlyStopping here.
- ModelCheckpoint callback is used with training using model.fit() to save the weights at some interval, so the weights can be loaded later to continue the training from the state saved.
Read more about ModelCheckpoint parameters here.
# use early stopping to exit training if validation loss is not decreasing even after certain epochs earlystopping = EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=20) # save the model with least validation loss checkpointer = ModelCheckpoint(filepath="classifier-resnet-weights.hdf5", verbose=1, save_best_only=True)
- Now, you train the model and give the callbacks defined above in the parameter.
model.fit(train_generator, steps_per_epoch= train_generator.n // 16, epochs = 1, validation_data= valid_generator, validation_steps= valid_generator.n // 16, callbacks=[checkpointer, earlystopping])
- Predict and convert the predicted data into a list.
# make prediction test_predict = model.predict(test_generator, steps = test_generator.n // 16, verbose =1) # Obtain the predicted class from the model prediction predict = [] for i in test_predict: predict.append(str(np.argmax(i))) predict = np.asarray(predict)
- Measure the accuracy of the model.
# Obtain the accuracy of the model from sklearn.metrics import accuracy_score accuracy = accuracy_score(original, predict) accuracy

- Print the classification report.
from sklearn.metrics import classification_report report = classification_report(original, predict, labels = [0,1]) print(report)
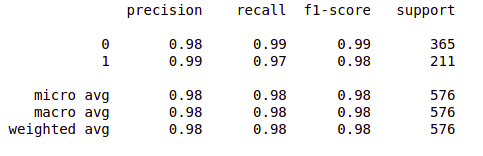
So far so good!
Now, you can pat yourself as you have just completed the first part i.e., brain tumor detection. Now, we will head towards brain tumor segmentation in the second part of this series.
Brain Tumor Detection and Localization using Deep Learning: Part 2
Being a Data Science enthusiast, I write similar articles related to Machine Learning, Deep Learning, Computer Vision, and many more. Follow my medium account: https://medium.com/@mahishapatel to get updates about my articles.
Also, I’d love to connect with you on Linkedin: https://www.linkedin.com/in/mahisha-patel/
Your suggestions and doubts are welcomed here in the comment section. Thank you for reading my article!







Hi, Thank you The link to Part 2 of your article is broken.
Hello, I couldn't find the dataset that you shared a link. It gives a different dataset from yours.
where is the original data set while prediction