This article was published as a part of the Data Science Blogathon
What are Genetic Algorithms?
Genetic Algorithms are search algorithms inspired by Darwin’s Theory of Evolution in nature.
By simulating the process of natural selection, reproduction and mutation, the genetic algorithms can produce high-quality solutions for various problems including search and optimization.
By the effective use of the Theory of Evolution genetic algorithms are able to surmount problems faced by traditional algorithms.
According to Darwin’s theory of evolution, an evolution maintains a population of individuals that vary from each other (variation). Those who are better adapted to their environment have a greater chance of surviving, breeding, and passing their traits to the next generation (survival of the fittest).
How do Genetic Algorithms work?
Before getting into the working of a Genetic Algorithm let us dive into the basic terminologies of Genetic Algorithms.
Chromosome/Individual
A chromosome is a collection of genes. For example, a chromosome can be represented as a binary string where each bit is a gene.
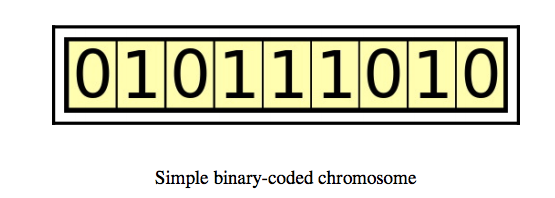
Image Source: Hands-On Genetic Algorithm with Python, Eyal Wirsansky
Population
Since an individual is represented as a chromosome, a population is a collection of such chromosomes.
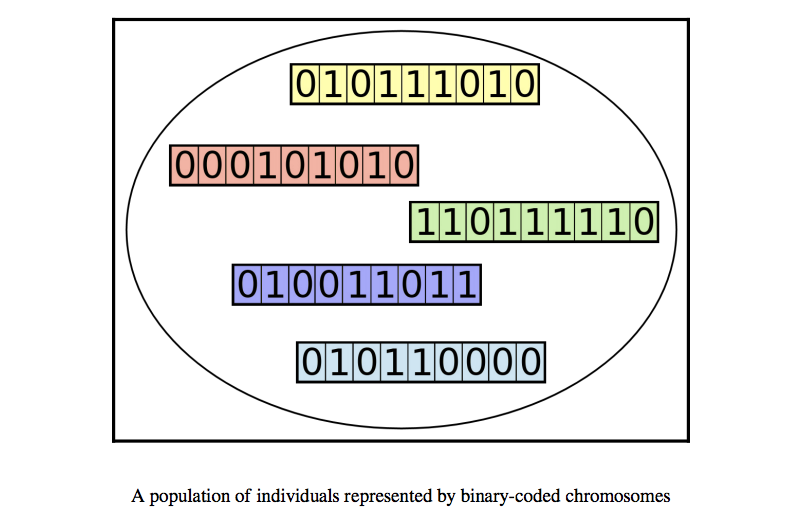
Image Source: Hands-On Genetic Algorithm with Python, Eyal Wirsansky
Fitness Function
Individuals who achieve a better fitness score represent better solutions and are more likely to be chosen to crossover and passed on to the next generation.
Selection
Types of selection methods available,
- Roulette wheel selection
- Tournament selection
- Rank-based selection
Crossover
Generally, two individuals are chosen from the current generation and their genes are interchanged between two individuals to create a new individual representing the offspring. This process is also called mating or crossover.
Types of crossover methods available,
- One point crossover
- Two-point crossover
- Uniform crossover
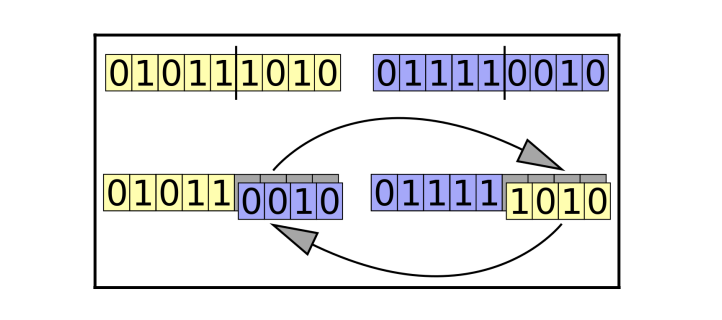
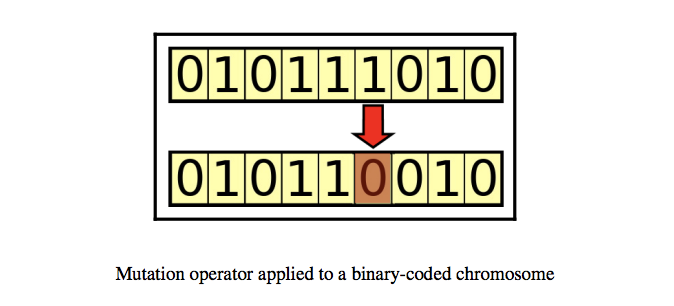
Mutation
The mutation is a random change in a chromosome to introduce new patterns to a chromosome. For example, flipping a bit in a binary string.
- Flip bit mutation
- Gaussian mutation
- Swap mutation
General workflow of a simple genetic algorithm
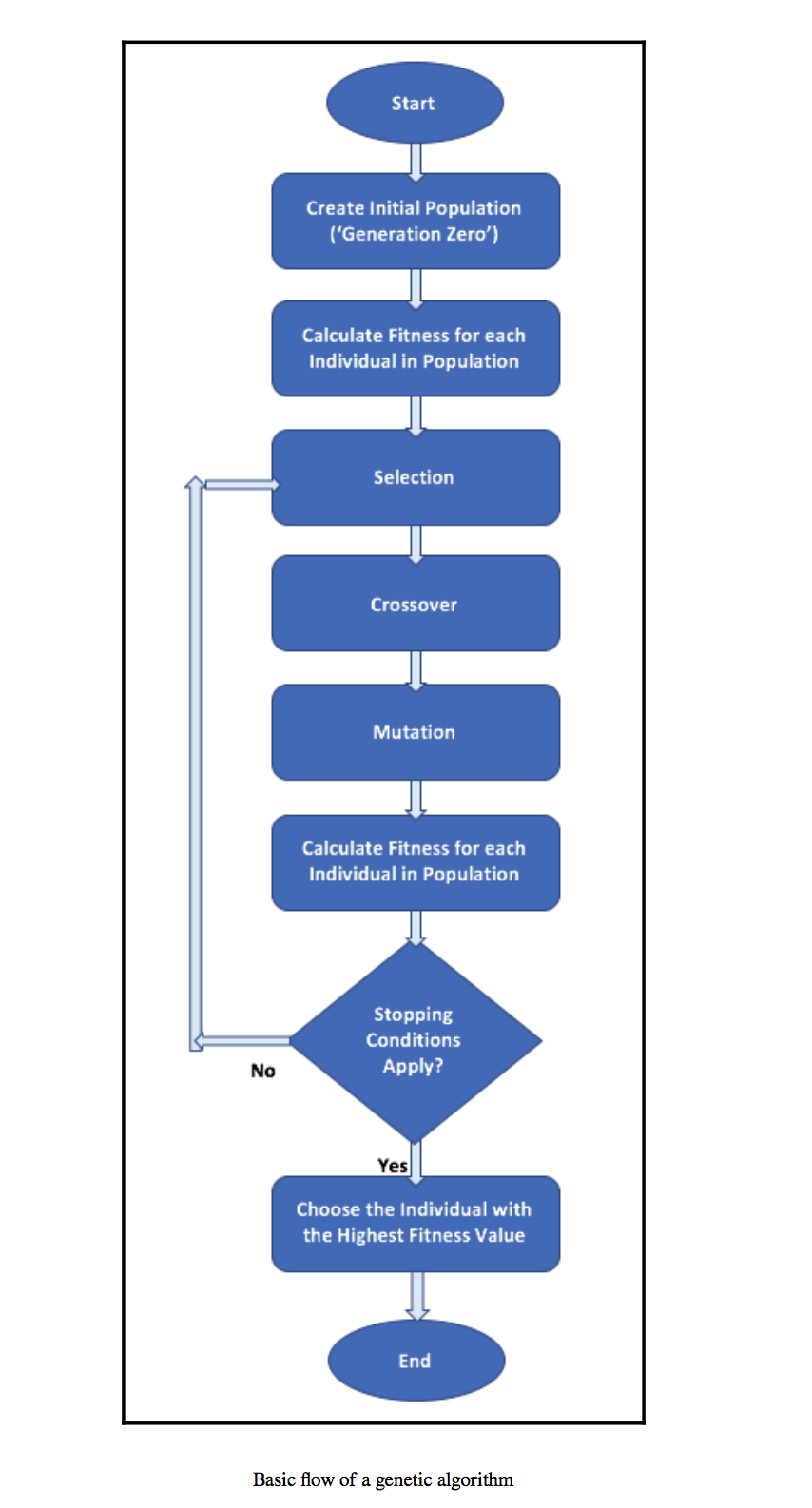
Image Source: Hands-On Genetic Algorithms with Python, Eyal Wirsansky
How do genetic algorithms differ from traditional algorithms?
A search space is a set of all possible solutions to the problem. Traditional Algorithms maintain only one set in a search space whereas Genetic Algorithms use several sets in a search space (Feature selection using R.F.E vs. Genetic Algorithms).
Traditional Algorithms require more information to perform a search whereas Genetic Algorithms just require one objective function to calculate the fitness of an individual.
Traditional Algorithms cannot work in parallel whereas Genetic Algorithms can work in parallel (calculating the fitness of the individuals are independent).
One big difference in Genetic Algorithms is that instead of operating directly on candidate solutions, genetic algorithms operate on their representations (or coding), often referred to as chromosomes.
Traditional Algorithms can only produce one solution in the end whereas in Genetic Algorithms multiple optimal solutions can be obtained from different generations.
Traditional Algorithms are not more likely to produce global optimal solutions, Genetic Algorithms are not guaranteed to produce global optimal solutions as well but are more likely to produce global optimal solutions because of the genetic operators like crossover and mutation.
Traditional algorithms are deterministic in nature whereas genetic algorithms are probabilistic and stochastic in nature.
Real-world problems are multi-modal (contains multiple locally optimal solutions), the traditional algorithms don’t handle well these problems whereas Genetic Algorithms, with the right parameter setting, can handle these problems very well because of the large solution space.
Advantages of Genetic Algorithms
Parallelism
Global optimization
A larger set of solution space
Requires less information
Provides multiple optimal solutions
Probabilistic in nature
Genetic representations using chromosomes
Disadvantages of Genetic Algorithms
The need for special definitions
Hyper-parameter tuning
Computational complexity
Use-cases of Genetic Algorithms in Machine Learning
Feature Selection
Model Hyper-parameter Tuning
Machine Learning Pipeline Optimization
TPOT(Tree-Based Pipeline Optimization) is an Auto-ML framework that utilizes genetic algorithms to optimize machine learning pipelines using the Genetic Algorithm framework called DEAP (Distributed Evolutionary Algorithms in Python).
Feature Selection using Genetic Algorithms
Using Genetic Algorithms, let us perform feature selection on a wine dataset available in sklearn.
import random import pandas as pd from sklearn.base import clone from deap.algorithms import eaSimple from deap import base, creator, tools from sklearn.datasets import load_wine from sklearn.metrics import accuracy_score from sklearn.ensemble import RandomForestClassifier from sklearn.model_selection import train_test_split Python Code:
import pandas as pd
from sklearn.datasets import load_wine
data = load_wine()
features = pd.DataFrame(
data.data, columns=data.feature_names)
print(features.head())
target = pd.Series(data.target)
print(target.head())# split the data into train and test x_train, x_test, y_train, y_test = train_test_split( features, target, test_size=0.2, stratify=target ) model = RandomForestClassifier()
Split the data into features and target and then split the features and target for training and testing.
def compute_fitness_score(individual):
"""
Select the features from the individual, train
and compute the accuracy_score.
Example:
individual = [0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1]
The 1 represents the presence of features and
0 represents the absence of features
"""
column_support = pd.Series(individual).astype(bool)
global x_train, y_train, x_test, y_test, model
x_train_ = x_train[x_train.columns[column_support]]
x_test_ = x_test[x_test.columns[column_support]]
model.fit(x_train_, y_train)
y_pred = model.predict(x_test_)
score = accuracy_score(y_test, y_pred)
return score,
Define the fitness score. compute_fitness_score takes in an individual as an input, for example, let us consider the following individual [0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1], in this list 1 represents the presence of that particular feature and 0 represents the absence of that feature. The columns are selected according to the individual and then the model is fitted and the accuracy score is calculated. This accuracy score is the fitness score of an individual.
n_genes = x_train.shape[1]
n_generations = 10
n_population = 10
crossover_probability = 0.6
mutation_probability = 0.2
def setup_toolbox():
# container for individuals
creator.create('FitnessMax', base.Fitness, weights=(1.0,))
creator.create('Individual', list, fitness=creator.FitnessMax)
toolbox = base.Toolbox()
toolbox.register(
'individual_generator_function',
random.randint, 0, 1
)
# method to populate individual
toolbox.register(
'individual_generator',
tools.initRepeat,
creator.Individual,
toolbox.individual_generator_function,
n_genes
)
# method to create population
toolbox.register(
'population_generator',
tools.initRepeat,
list,
toolbox.individual_generator
)
# fitness calculation
toolbox.register(
'evaluate', compute_fitness_score
)
# selection
toolbox.register(
'select', tools.selTournament, tournsize=3
)
# crossover
toolbox.register('mate', tools.cxOnePoint)
# mutation
toolbox.register(
'mutate',
tools.mutFlipBit,
indpb=mutation_probability
)
return toolbox
n_genes represent the number of genes in an individual which is equal to the number of features, n_generations represent the number of generations which is equal to 10 and so is n_population which represents the number of population. The cross-over probability is set to 0.6 and the mutation probability is set to 0.2.
Toolbox is a class in deap framework that provides us required utilities to define individual, population, and much more.
Steps done in setup_toolbox method are,
- First, we define an Individual which has a fitness score associated with it
- Next, we define a function that generates genes of an individual, in our case 0 or 1 in an individual
- We define a function that generates individuals of a population, for example, [0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1] is an individual in a population.
- Next, we define a function that generates a population, for example, [[0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 1],[0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1]] is a population of size 2.
- Next, we define a function for evaluation, in our case we use the function compute_fitness_score.
- We chose the tournament selection method for the selection process, one point cross-over for the cross-over process, and flip bit mutation for the mutation process.
# setup deap toolbox
toolbox = setup_toolbox()
# create a population
population = toolbox.population_generator(n_population)
# A simple evolutionary algorithm
final_population, logbook = eaSimple(
population,
toolbox,
crossover_probability,
mutation_probability,
n_generations
)
We first generate a population of individuals of size 10. Then we call eaSimple method in deap which performs a simple genetic algorithm.
The arguments of eaSimple function,
- The first argument is the population
- toolbox is the instance of deap’s Toolbox class
- The cross-over probability denotes the probability of a pair of individuals to be crossed over
- The mutation probability denotes the probability of an individual to be mutated
- n_generations denotes the number of generations to evolve.
The eaSimple algorithm works as follows,
- The fitness of the individuals in a population is calculated
- Select the fittest individual using the tournament selection method
- Cross-over the individuals using one point cross-over method
- Mutate the individuals using flip bit mutation
- Calculate the fitness of the individuals again
- Follow steps 2 through 5 until the desired number of generations is reached
In this way, you can perform feature selection using a Genetic algorithm.
References
Thank you!
The media shown in this article are not owned by Analytics Vidhya and are used at the Author’s discretion.


