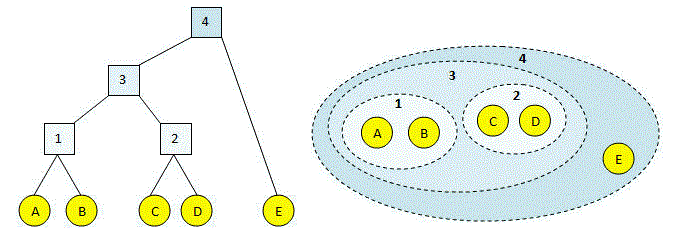
As we all know, single-link Hierarchical clustering begins by treating each observation as an individual cluster and then iteratively merges clusters until all the data points form a single cluster. Transitioning to the use of dendrograms to represent hierarchical clustering results, clusters merge based on the distance between them.
Various types of linkages, such as single linkage, complete linkage, and average linkage, utilize different methods to calculate the distance between clusters.
Learning Objectives:
- Understand the concept and purpose of Linear Discriminant Analysis (LDA)
- Learn how LDA performs dimensionality reduction and classification
- Grasp the mathematical principles behind Fisher’s Linear Discriminant
- Explore LDA implementation and applications using Python
This article was published as a part of the Data Science Blogathon
Table of contents
Linkage Criteria
It determines the distance between sets of observations as a function of the pairwise distance between observations.
- Furthermore, in Single Linkage Clustering, the distance between two clusters is the minimum distance between members of the two clusters.
- Additionally, in Complete Linkage, the distance between two clusters is the maximum distance between members of the two clusters.
- Moreover, in Average Linkage, the distance between two clusters is the average of all distances between members of the two clusters.
- Finally, in Centroid Linkage, the distance between two clusters is the distance between their centroids.

Similarly, in this article, we aim to understand the Clustering process using the Single Linkage Clustering Method.
Clustering Using Single Linkage
Begin with importing necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import scipy.cluster.hierarchy as shc
from scipy.spatial.distance import squareform, pdistLet us create toy data using numpy.random.random_sample
a = np.random.random_sample(size = 5)
b = np.random.random_sample(size = 5)Once we generate the random data points, we will create a pandas data frame.
point = ['P1','P2','P3','P4','P5']
data = pd.DataFrame({'Point':point, 'a':np.round(a,2), 'b':np.round(b,2)})
data = data.set_index('Point')
data
A glance at our toy data. Looks clean. Let us jump into the clustering steps.
Step1: Visualize the data using a Scatter Plot
plt.figure(figsize=(8,5))
plt.scatter(data['a'], data['b'], c='r', marker='*')
plt.xlabel('Column a')
plt.ylabel('column b')
plt.title('Scatter Plot of x and y')for j in data.itertuples():
plt.annotate(j.Index, (j.a, j.b), fontsize=15)
Step2: Calculating the distance matrix in Euclidean method using pdist
dist = pd.DataFrame(squareform(pdist(data[[‘a’, ‘b’]]), ‘euclidean’), columns=data.index.values, index=data.index.values)For our convenience, we will be considering only the lower bound values of the matrix as shown below. Specifically, the lower bound values represent the minimum distance between any two points in the dataset.

Step 3: Look for the least distance and merge those into a cluster

We see the points P3, P4 has the least distance “0.30232”. So we will first merge those into a cluster.
Step 4: Re-compute the distance matrix after forming a cluster
Update the Distance Between
Cluster (P3,P4) to P1
= Min(dist(P3,P4), P1)) -> Min(dist(P3,P1),dist(P4,P1))
= Min(0.59304, 0.46098)
= 0.46098Cluster (P3,P4) to P2
= Min(dist(P3,P4), P2) -> Min(dist(P3,P2),dist(P4,P2))
= Min(0.77369, 0.61612)
= 0.61612And Cluster (P3,P4) to P5
= Min(dist(P3,P4), P5) -> Min(dist(P3,P5),dist(P4,P5))
= Min(0.45222, 0.35847)
= 0.35847
Repeat steps 3 and 4 until you are left with one single cluster.
After re-computing the distance matrix, we need to again look for the least distance to make a cluster.
Subsequently, we repeat this process until we have clustered all observations.

We see the points P2, P5 has the least distance “0.32388”. So we will group those into a cluster and recompute the distance matrix.
Update the distance between the cluster (P2,P5) to P1
= Min(dist((P2,P5),P1)) -> Min(dist(P2,P1), dist(P5, P1))
= Min(1.04139, 0.81841)
= 0.81841Update the distance between the cluster (P2,P5) to (P3,P4)
= Min(dist((P2,P5), (P3,P4))) -> = Min(dist(P2,(P3,P4)), dist(P5,(P3,P4)))
= Min(dist(0.61612, 0.35847))
= 0.35847
After recomputing the distance matrix, we need to again look for the least distance.

The cluster (P2,P5) has the least distance with the cluster (P3, P4) “0.35847”. So we will cluster them together.
Update the distance between the cluster (P3,P4, P2,P5) to P1
= Min(dist(((P3,P4),(P2,P5)), P1))
= Min(0.46098, 0.81841)
= 0.46098
We have completed obtaining a single cluster.
Theoretically, the clustering steps proceed as follows:
- Transitioning to active voice: P3 and P4 points merge due to their least distance.
- Next, P2 and P5 points merge as they exhibit the least distance.
- Subsequently, we cluster the pairs (P3, P4) and (P2, P5).
- Finally, we merge the cluster (P3, P4, P2, P5) with the datapoint P1.
We can visualize the same using a dendrogram
plt.figure(figsize=(12,5))
plt.title("Dendrogram with Single inkage")
dend = shc.dendrogram(shc.linkage(data[['a', 'b']], method='single'), labels=data.index)
The length of the vertical lines in the dendrogram shows the distance. For example, the distance between the points P2, P5 is 0.32388.
The step-by-step clustering that we did is the same as the dendrogram
Conclusion
Single linkage clustering involves several key steps. Initially, after visualizing the data and calculating a distance matrix, clusters are formed based on the shortest distances. Once each cluster is formed, the algorithm updates the distance matrix to incorporate new distances. Throughout this iterative process, all data points are clustered until revealing patterns in the data. Therefore, it’s a simple, intuitive method that can uncover hidden structures in the data.
Key Takeaways:
- Firstly, LDA maximizes between-class scatter while minimizing within-class scatter
- Moreover, it assumes Gaussian distribution and identical covariance matrices for classes
- Additionally, LDA can be extended for multi-class problems and addresses some limitations of logistic regression
- Finally, regularization techniques help overcome LDA’s small sample size problem
Frequently Asked Questions
A. Single link hierarchical clustering, also known as single linkage clustering, merges clusters based on the closest pair of points between them. It forms clusters where the smallest pairwise distance between points is minimized.
A. Hierarchical clustering includes agglomerative and divisive methods. Agglomerative clustering, which includes single link clustering, merges data points into progressively larger clusters. Divisive clustering, in contrast, starts with one cluster and splits it into smaller ones.
A. The agglomerative hierarchical clustering method known as single link uses the nearest neighbor approach to merge clusters. Specifically, it connects clusters based on the shortest distance between any two points in different clusters.
A. The linkage method in hierarchical clustering problem determines how distances between clusters are calculated during the merging process. Common linkage methods include single (or nearest neighbor), complete (or farthest neighbor), average, and Ward’s method.






Nice Explanation. I really appreciate all of your work