This article was published as a part of the Data Science Blogathon
Overview
This article aims to help those who are facing the same issues as me: working with Python but missing the OpenAir package of R.
Since I started with Python, I’ve never found a way of exploring pollutant datasets the way OpenAir does. So, using the Seaborn library, I tried to develop two of the visualizations I liked the most in the R package.
Then I’ll show both functions created and I hope it can help you.
Introduction
Recently, much has been discussed about air pollution and its consequences on the environment. These discussions always gain prominence when some of their consequences haunt the world and leave us wondering what will be of future generations.
Maybe greenhouse gases and the consequent global warming are the most well-known names for most of the population. However, other air pollutants cause many harmful effects to the population and end up not being well regulated in many regions. These are the cases of particulate matter and nitrogen oxides, for example. In large cities, these pollutants are usually emitted on a large scale by diesel vehicles that lack proper maintenance.
For a more assertive assessment of the state of pollutant concentrations in a given region, an in-depth study of the data is necessary. For that, a couple of years ago I started using the OpenAir package of R, which by the way is excellent. However, over the last few months, I’ve started using Python and I’ve come to like it. But what I miss the most about R is OpenAir.
With that in mind, I decided to develop two functions in Python, for visualizing pollutants datasets, inspired by OpenAir of R.
So, in this article, I’ll present these functions and I’ll detail how they were developed.
It’s important to make it clear that I’m not going to deal with the analysis of local pollutants here, but rather use the data as an example to illustrate our analysis tools. By the way, I used a dataset from: https://qualar.cetesb.sp.gov.br/qualar/home.do, which is the agency responsible for regulating environmental issues in the state of Sao Paulo.
What are the functions I’ve developed?
The first one is a set of plots, which I called time_variation, showing the concentration of the pollutant by the hour, month, and day of the week, like the image below.
The second one is a calendar heatmap (calendar_dist), which points the worst days relating to pollutant concentrations:
Time Variation
- The first part, 3 plots (concentrations by the hour, month, and day of the week)
First things first. To kick off our function, we set the input variables (df, pollutant, ylabel, hue=None ), and also, we have to import the libraries.
def time_variation(df, pollutant, ylabel, hue=None):
# importing all the libraries we'll need
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
As we’re going to use int values representing months and days of the week to create the charts, we need lists of labels to replace these int values in the plots.
#setting xticklabels
week = ['mon','tue','wed','thu','fri','sat','sun']
months = ['jan','feb','mar','apr','may','jun','jul','aug','sep','oct','nov','dec']
To make our function work fine, we need to be sure the data in feature date is really in date format. So:
# converting feature date to datetime and dropping Nan values
df['date'] = pd.to_datetime(df['date'])
if hue!=None:
df = df[['date', pollutant, hue]].dropna() # dropping rows of nan vals of the chosen pollutant
else:
df = df[['date', pollutant]].dropna()
Now, we only need to plot. But wait, first let’s set the subplots. There will always be 3 subplots, representing concentrations by the hour, month, and day of the week.
# Creating graphs of pollutant concentrations by hour, month and day of week
fig,axes = plt.subplots(1, 3,sharex=False, figsize=(16,4)) #creating subplots, side by side
fig.tight_layout(pad=-2) # makeing plots get closer
sns.set_style('whitegrid')
After that, plotting the graphs is very simple. In the example below, I show the lineplot of concentration by the hour. The others are very similar, we just have to change hours by month or dayofweek.
# concentration vs hour
axes[0] = sns.lineplot(ax=axes[0],data=df,
x=df['date'].dt.hour,
y=pollutant,
color='red',
linewidth=1.5,
hue=hue,
palette="hls")
axes[0].set_xticklabels(axes[0].get_xticks(), fontsize=13)
axes[0].set_yticklabels(axes[0].get_yticks(), fontsize=13)
axes[0].set_xlabel('hour', fontsize=15)
axes[0].set_ylabel(ylabel, fontsize=15)
axes[0].legend().set_title('')
# concentration vs month
axes[1] = sns.lineplot(ax=axes[1],
data=df,
x=df['date'].dt.month,
y=pollutant,
color='red',
linewidth=1.5,
hue=hue,
palette="hls")
axes[1].set_xticks(np.arange(1, 13, 1))
axes[1].set_xticklabels(months, fontsize=13)
axes[1].set_yticklabels('')
axes[1].set_xlabel('month', fontsize=15)
axes[1].set_ylabel('')
axes[1].legend().set_title('')
# concentration vs day of week
axes[2] = sns.lineplot(ax=axes[2],
data=df,
x=df['date'].dt.dayofweek,
y=pollutant,
color='red',
linewidth=1.5,
hue=hue,
palette="hls")
axes[2].set_xticks(np.arange(0, 7, 1))
axes[2].set_xticklabels(week, fontsize=13)
axes[2].set_yticklabels('')
axes[2].set_xlabel('day of week', fontsize=15)
axes[2].set_ylabel('')
axes[2].legend().set_title('')
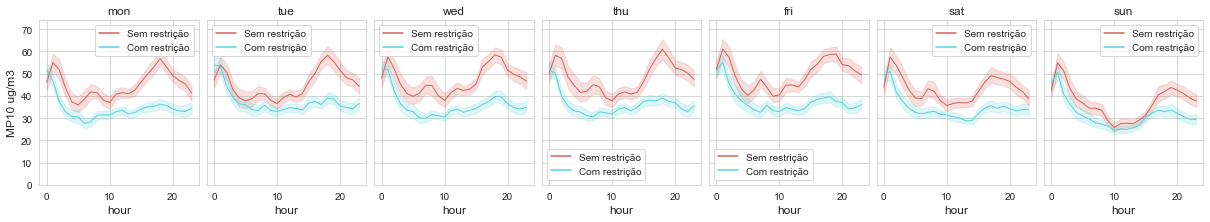
- Plotting concentrations by the hour, by days of the week
In the second part of our Time Variation, we want to check if there is any difference between mean pollutant concentrations by the hour and by days of the week. This is a great tool to use for checking the seasonality of a pollutant during a week. In the example above, it’s clear that the pollutant concentrations decrease on weekends, which can be related to the reduction of car traffic on these days.
Plotting it is as simple as the one before. As the subplots will always have the same numbers of rows and columns, we have:
# creating graphs of concentration by hour by specific day of week
fig2,axes2 = plt.subplots(1, 7,sharex=True, figsize=(16,2)) #subplots for each day of week
fig2.tight_layout(pad=-2)
Now, we just have to insert the graphs on the subplots using Seaborn lineplot:
#Plots
if hue!=None:
pollutant_max=max(df.groupby([df['date'].dt.hour, hue])[pollutant].mean())*1.3 #setting the lim of y due to max mean
else:
pollutant_max=max(df.groupby(df['date'].dt.hour)[pollutant].mean())*1.3
for i in range(7):
axes2[i] = sns.lineplot(ax=axes2[i],data=df,
x=df[df.date.dt.dayofweek==i]['date'].dt.hour,
y=pollutant,
color='red',
linewidth=1,
hue=hue,
palette="hls")
axes2[i].set_xlabel('hour', fontsize=12)
if i == 0:
axes2[i].set_ylabel(ylabel, fontsize=12)
else:
axes2[i].set_ylabel('')
axes2[i].set_yticklabels('')
axes2[i].set_ylim(0, pollutant_max)
axes2[i].set_title(week[i])
axes2[i].legend().set_title('')
return fig.show(), fig2.show()
And, that’s all for our Time Variation function!
Using another example, we got:
time_variation(df_evandro_restricao, 'MP10', 'MP10 ug/m3', 'restricao')
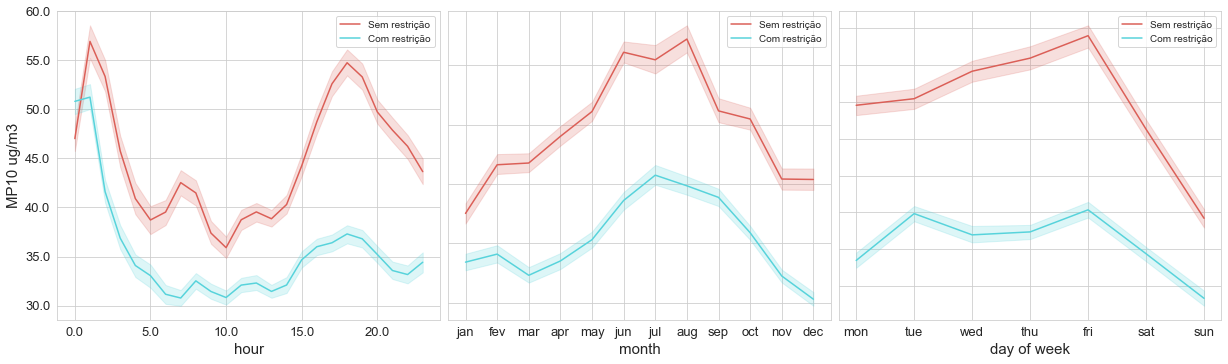
This image above represents concentrations of PM10 divided into two different portions of time, each one having a different limit for pollutant emissions.
Calendar Distribution (calendar_dist)
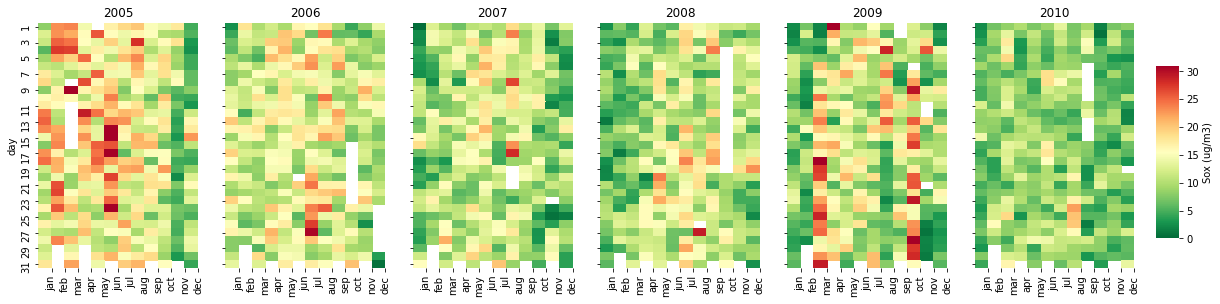
A calendar distribution is a powerful tool to construct a nice visualization of pollutants concentrations along the days and years. To build this function, we need to set the inputs, libraries, a list of months and create new columns (year, month, and day).
def calendar_dist(df, pollutant, cbar_label, vmin=0, vmax=100):
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
import math
# Setting feature date into datetime and creating columns year, month and day
df.date = pd.to_datetime(df.date)
df['year'] = df['date'].dt.year
df['month'] = df['date'].dt.month
df['day'] = df['date'].dt.day
months = ['jan','feb','mar','apr','may','jun','jul','aug','sep','oct','nov','dec']
Note that this time I’ve imported the math library too. It’ll be important later.
Since our goal is to make a calendar distribution for each year of our dataset, I need to count the number of years we have to build the subplots. Also, I noticed that the maximum subplots per row, that keep visualization still nice, is 6. So, we have:
years = df.year.unique()
n_years = len(df.year.unique())
sub_row = int(math.ceil(n_years/6))
high = sub_row*4 # high of the colorbar
Now, we just have to construct the calendars, using the function heatmap of Seaborn. But wait! Before start using the heatmap, we have to set our subplots and dataset. First of all, setting subplots will depend on the number of years we have. We have three different cases here: having just one year, having > 1 and <= 6 years, and more and 6 years. Note that for n_years==1 we don’t need to create subplots, since it’ll have just one heatmap. For each case, we build subplots by:
if n_years 1:
fig,axes = plt.subplots(1, n_years,sharex='col', sharey='row',figsize=(16,4))
fig.tight_layout(h_pad=1.0, w_pad=0.5) # makeing plots get closer
elif n_years==1:
elif n_years==1:
# here we don't need to create subplots
else:
else:
fig,axes = plt.subplots(sub_row, 6,sharex='col', figsize=(16,high), sharey = 'row') #creating subplots, side by side
fig.tight_layout(h_pad=1.0, w_pad=-1.5) # makeing plots get closer
As you can see, the first case (n_years 1) is simple. As we have just one row, the number of subplots will be determined by n_years. The second one is even easier since we don’t need a subplot. The third one has a number of rows determined by sub_row, which is the next integer value from the division of n_years by 6.
Since the heatmap function uses the interception value of a column and a row indexes, we do the following for each calendar:
# creating a table of relation between month and day, in specific year, giving pollutant conc. mean as result
df_pivot = df[df['year'].isin([years[i]])].groupby(['month', 'day'])[pollutant].mean()
df_pivot = pd.DataFrame(df_pivot).reset_index()
df_pivot = df_pivot.pivot('month','day',pollutant)
Now, we can finally use heatmap and build our calendars! For the first case, we just have to use:
if n_years 1:
fig,axes = plt.subplots(1, n_years,sharex='col', sharey='row',figsize=(16,4))
fig.tight_layout(h_pad=1.0, w_pad=0.5) # makeing plots get closer
# creating a table of relation between month and day, in specific year, giving pollutant conc. mean as result
df_pivot = df[df['year'].isin([years[i]])].groupby(['month', 'day'])[pollutant].mean()
df_pivot = pd.DataFrame(df_pivot).reset_index()
df_pivot = df_pivot.pivot('month','day',pollutant)
cbar_ax = fig.add_axes([1, .2, .02, sub_row*0.3])
if i==n_years-1:
axes[i] = sns.heatmap(ax = axes[i], data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar_ax = cbar_ax,
cbar_kws={'label': cbar_label},
cbar=True)
else:
axes[i] = sns.heatmap(ax = axes[i], data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar=False)
axes[i].set_xlabel('')
axes[i].set_title(years[i])
axes[i].set_xticks(np.arange(1, 13, 1))
axes[i].set_xticklabels(months, rotation=90)
if i==0:
axes[i].set_ylabel('day')
else:
axes[i].set_ylabel('')
for i in range(n_years):
Note that in this case, I just built a conditional statement inside the loop to make the colorbar be set only once.
The second case is easier, we just need to plot the new dataframe (df_pivot) using the heatmap:
elif n_years==1:
df_pivot = df[df['year'].isin([years[0]])].groupby(['month', 'day'])[pollutant].mean()
df_pivot = pd.DataFrame(df_pivot).reset_index()
df_pivot=df_pivot.pivot('month','day',pollutant)
ax = sns.heatmap(data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar_kws={'label': cbar_label},
cbar=True)
ax.set_xticklabels(months, rotation=90)
The third case is a little more complex because I had to divide it into the other three cases, depending on which row of subplots the loop was. So, let’s take a look:
else:
fig,axes = plt.subplots(sub_row, 6,sharex='col', figsize=(16,high), sharey = 'row') #creating subplots, side by side
fig.tight_layout(h_pad=1.0, w_pad=-1.5) # makeing plots get closer
for i in range(n_years):
df_pivot = df[df['year'].isin([years[i]])].groupby(['month', 'day'])[pollutant].mean()
df_pivot = pd.DataFrame(df_pivot).reset_index()
df_pivot = df_pivot.pivot('month','day',pollutant)
if i<6: # for the first row
k=0
axes[k,i] = sns.heatmap(ax = axes[k,i], data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar=False)
axes[k,i].set_xlabel('')
axes[k,i].set_title(years[i])
if i==0:
axes[k,i].set_ylabel('day')
else:
axes[k,i].set_ylabel('')
elif i%6==0:
k=int(i/6)
j=int(i%6)
if i==n_years-1:
axes[k,j] = sns.heatmap(ax = axes[k,j], data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar_ax = cbar_ax,
cbar_kws={'label': cbar_label},
cbar=True)
else:
axes[k,j] = sns.heatmap(ax = axes[k,j], data=df_pivot.T,
vmin=vmin,
vmax=vmax,
cmap="RdYlGn_r",
cbar=False)
axes[k,j].set_xlabel('')
axes[k,j].set_title(years[i])
axes[k,j].set_xticklabels(months, rotation=90)
else:
k=int(math.ceil(i/6))-1
j=i%6
if i==n_years-1:
cbar_ax = fig.add_axes([1, .2, .02, sub_row*0.3])
axes[k,j] = sns.heatmap(ax = axes[k,j], data=df_pivot.T,
vmin=0,
vmax=max(df_pivot),
cmap="RdYlGn_r",
cbar_ax = cbar_ax,
cbar_kws={'label': cbar_label},
cbar=True)
else:
axes[k,j] = sns.heatmap(ax = axes[k,j], data=df_pivot.T,
vmin=0,
vmax=max(df_pivot),
cmap="RdYlGn_r",
cbar=False)
axes[k,j].set_xlabel('')
axes[k,j].set_title(years[i])
axes[k,j].set_xticklabels(months, rotation=90)
if j==0:
axes[k,j].set_ylabel('day')
else:
axes[k,j].set_ylabel('')
In this case, I had to set a new variable ‘k’ which is representing the rows of our subplots, while ‘i’ is being used in our list of years.
It’s important to note that I didn’t set the colorbar for all heatmaps, otherwise our figure would become very polluted. We just need to set cbar_ax once, for each case. Also using cbar_ax makes the heatmaps be the same size, which doesn’t happen with cbar=True.
One thing I didn’t mention but you may have noticed is that this function always creates the number of subplots multiple of 6. So, after setting our plots, it may have left some axes of subplots empty. To fix that, we do the following:
if n_years%6 != 0 and n_years>6:
rest = 6-n_years%6
for i in range(rest):
axes[1,5-i].axis('off')
So, here’s an example of the usage of this function:
calendar_dist(df,'MP10','MP10 (ug/m3)', vmax=100)
Conclusion
So I’ve just shown these two powerful functions for analyzing and visualizing pollutants concentrations along with a specific range of dates.
I can say that time_variation and calendar_dist are very inspired by the functions of OpenAir from R. Although both are used to visualize trends and seasonality of pollutants concentrations, each one has specific usability.
I hope this article can help many others who are working on this topic of research, and also I hope you enjoy it as I did.
If you have any questions, please leave your comment or contact me through my Linkedin profile:



