This article was published as a part of the Data Science Blogathon
Overview
In the current scenario, the Data science field is dominated by Python/R but there is another competition added not so long ago, Julia! which we will be exploring in this guide. The famous quote (motto) of Julia is –
Looks like Python, runs like C
We know that python is used for a wide range of tasks. Julia, on the other hand, was primarily developed to perform scientific computation, machine learning, and statistical tasks.
Since Julia was explicitly made for high-level statistical work and scientific computations, it has several benefits over Python. In linear algebra, for example, “vanilla” (raw) Julia performs better than “vanilla” (raw) Python. This is primarily because, unlike Julia, Python doesn’t support all equations and the matrices used in machine learning.
While Python is a great language with its library Numpy, Julia completely outperforms it when it comes to non-package experience, with Julia being more catered towards machine learning tasks and computations.
Table of contents
- Introduction
- Installation
- Arrays
- Cuda Arrays
- Automatic Differentiation
- Training a Classifier
- Conclusion
So, let’s get started!
Introduction
This guide is to get you started with the mechanics of Flux, to start building models right away. While this is loosely based on a tutorial by Pytorch, it will cover all the areas necessary. It introduces basic Julia programming, as well Zygote, a source-to-source automatic differentiation (AD) framework in Julia. Using all these tools, we will build a simple neural network and in the end a CNN which we will train to classify between 10 classes.
What is Flux in Julia?
Flux is an open-source machine-learning software library written completely in Julia. A stable release which we will be using is v0.12.4. As we would have expected, it has a layer-on layer stacking-based interface for simple models with strong support on interoperability with other packages of Julia, instead of having a monolithic design. For example, if we need GPU support we can get it directly via the implementation of CuArrays. This is in complete contrast to other frameworks in Julia which are implemented in different languages but bound with Julia such as Tensorflow (Julia Package) and thus are more or less limited by the functionality present in their implementation.
Installation of Julia
Before we move further, if you don’t have Julia installed in your system, it can be from its official site julialang.org .
To use Julia in Jupyter notebook like Python, we only need to add the IJulia package as follows and we can run Julia right from the jupyter notebook.
using Pkg
Pkg.add("IJulia")
We can use Julia as we used Python in Jupyter notebook for exploratory data analysis.
Arrays in Julia
Before moving on to the framework, we need to understand the basics of a deep learning framework. Arrays, CudaArrays, etc. In this section, I’ll explain the basics of the array in Julia.
The starting point for all of our models is the Array (also referred to as a Tensor for example in Pytorch and in other frameworks). It’s just a list of numbers of elements that can also be arranged in the form of a square (matrix). Let’s write an array
with just three elements.
x = [10,12,16]

Here’s a matrix – a square array with four elements.
x = [10 12; 13 14]
Since usually, we work with hundreds and thousands of arrays, we don’t always write them by hand. So, here’s how we can create a Matrix of 12×2= 24
elements, each a random number ranging from 0 to 1.
x = rand(12, 2)
rand is not just the only function that can create a random matrix (array) we can use different functions like ones, zeros, or randn. Try them out in the jupyter notebook to see what they do.
By default, Julia stores all the numbers in a high-precision format called Float64. In Machine Learning we often don’t need all those many digits, so we can configure Julia to decrease it to Float32, or if we need higher precision than 64 bits we can use BigFloat. Below is an example of a random matrix of 6×3 = 18 elements of BigFloat.
x = rand(BigFloat, 6, 3)

x = rand(Float32, 6, 3)
To count the number of elements in a matrix we can use the length function.
length(x)
Or, if we need the size we can check it more explicitly.
size(x)
We can do many sorts of algebraic operations on matrix, for example, we can add two matrices
x + x

Or subtract them.
x - x

Julia supports a feature called broadcasting, using the “.” syntax. The broadcast() is an inbuilt function in julia that is used to broadcast or apply the function f over the collections, arrays, or tuples. This makes it easy to apply a function to one or more arrays with a concise dot. syntax. For example – f.(a, b) means “apply f elementwise to a and b”. We can use this broadcasting in our matrix to add 1 element-wise in x.
x .+ 1
Finally, we have to use Matrix Multiplication more or less every time we use Machine Learning. Is super-easy to use with Julia.
W = randn(4, 10)
x = rand(10)
W * x
CUDA Arrays in Julia
CUDA functionality is provided separately by the CUDA package from Julia. If you have a GPU and CUDA available, you can run ] add CUDA in IJulia (jupyter notebook) to get it. Once you get the CUDA installed (compatible versions below julia 1.6) we can transfer our arrays into CUDA arrays (or in GPU) using cu function. It supports all the basic functionalities of an array but now works on GPU.
We are already aware of the parallelism power of GPU, the easiest method to get started is using Arrays. CUDA.jl (Julia Package) provides an array type, CuArray,
and many specialized array operations that execute efficiently on the
GPU hardware. In this section, I will briefly demonstrate the use of the CuArray type. Since we are exposing CUDA’s functionality by implementing existing Julia interfaces on the CuArray type, we should refer to the upstream Julia documentation for more in-depth information on these operations.
If you are running Julia >= 1.6 simply add Cuda as follows
import Pkg
Pkg.add("CUDA")
Import the library and covert matrix into CudaArrays. Now, these CuArrays will run on GPU which by default is much faster than Arrays and we barely had to do anything in it.
using CUDA
x = cu(rand(6, 3))
Flux.jl in Julia
Flux is a library or package in Julia specifically for machine learning. It comes with a vast range of functionalities that help us harness the full potential of Julia, without getting our hands messy (like auto-differentiation). We follow a few key principles in Flux.jl
- Doing the obvious –
Flux has relatively few explicit APIs for features like regularisation or embeddings which we need often. Instead, writing down the mathematical form will work –
and will be faster. - You could have written Flux from scratch – From LSTMs to GPU Kernels, it is a very straightforward Julia code. Whenever in doubt, one can always look to the documentation. If you need something different, you can easily write your own code in it.
- Integrates nicely with others – Flux works well with Julia libraries from Data Frames to Images (Images package) and even differential equations solver (another package in Julia for computation), so you can easily build complex data processing pipelines that integrate Flux models.
Installation
You can add Flux from using Julia’s package manager, by typing ] add Flux in the Julia prompt or use
import Pkg
Pkg.add("Flux")
Automatic Differentiation
Automatic differentiation (AD), also called algorithmic differentiation or simply “auto diff”, is used to calculate differentiation of functions. It is a family of techniques similar to backpropagation for efficiently evaluating derivatives of numeric functions expressed as a form of computer programs.
One probably has learned to differentiate functions in calculus classes but let’s recap it in Julia code.
f(x) = 4x^2 + 12x + 3 f(4)

In simpler cases like these, we can easily find the gradient by hand, for example in this it is 8x + 12. But it’s much faster and efficient to make the Flux do it for us!
using Flux: gradient
df(x) = gradient(f, x)[1]
df(4)
We can cross-check with few more inputs, to see if the gradient calculated by Flux is correct and is indeed 8x+12. We can do it multiple times and since the function we took was the C_2 function second derivative is just an integer 8.
ddf(x) = gradient(df, x)[1]
ddf(4)
As long as the mathematical functions we create in Julia are differentiable we can use auto differentiation of Flux to handle any code we throw at it, which includes recursion, loops, and even custom layers. For example, we can try to differentiate the Taylor series approximation of sin function.
mysin(x) = sum((-1)^k*x^(1+2k)/factorial(1+2k) for k in 0:6) x = 0.6 mysin(x), gradient(mysin, x)

sin(x), cos(x)
As we expected the derivative is numerically very close to the function cos(x) (which is sinx derivative).
What if instead of just taking a single number as input, we take arrays as inputs? This gets more interesting as we proceed further. Let’s take an example where we have a function that takes a matrix and two vectors.
myloss( W , b , x ) = sum(W * x .+ b) #calculating loss
W = randn(3, 5)
b = zeros(3)
x = rand(5)
gradient(myloss, W, b, x)Now we get gradients for each of the inputs W, b, and x, and these will come in very handy when we have to train our model. Since we know that machine learning models can contain hundreds or thousands of parameters, Flux here provides a slightly different method of writing gradient. Just like other deep learning frameworks, we mark our arrays with params to indicate that we want its gradients. W and b represent the weight and bias respectively.
using Flux: params
W = randn(3, 5)
b = zeros(3)
x = rand(5)
y(x) = sum(W * x .+ b)
grads = gradient(()->y(x), params([W, b]))
grads[W], grads[b]Using those parameters we can now get the gradients of W and b directly. It’s especially useful when we are working with layers. Think of the layer as a container for parameters. For example, the Dense function from Flux does familiar linear transform.
using Flux
m = Dense(10, 5)
x = rand(Float32, 10)To get parameters of any layer or model we can always simply use params from Flux.
params(m)
So even if our network has many many parameters we can easily calculate their gradient for all parameters.
x = rand(Float32, 10) #ran array m = Chain(Dense(10, 5, relu), Dense(5, 2), softmax) #creating a layer l(x) = sum(Flux.crossentropy(m(x), [0.5, 0.5])) #loss function grads = gradient(params(m)) do #calulating gradient l(x) endfor p in params(m) println(grads[p]) #printing parameters end
We don’t explicitly have to use layers but sometimes they can be very convenient for many simple kinds of models and faster iterations.
The next step would be to update the weights of the network and perform optimization using different algorithms. The first optimization algorithm which comes to mind is Gradient Descent because of its simplicity. We take the weights and steps using a learning rate which is hyper-param and the gradients. weights = weights – learning_rate x gradient.
using Flux.Optimise: update!, Descent
η = 0.1 #learning rate
for p in params(m)
update!(p, -η * grads[p]) #basic inplace update
endWhile the method we used above to update the param in place using gradients is valid, it can get way more complicated as the algorithms we use gets more involved in it. Here, Flux comes to the rescue with its prebuilt set of optimizers which makes our work way too easy. All we need to do is give the algorithm a learning rate and that’s it.
opt = Descent(0.01)So training a new network finally reduces down to iteration on the given dataset multiple times (epochs) and performing all the steps in order (given below in code). For the sake of simplicity and clarity, we do a quick implementation in Julia, let’s train a network that learns to predict 0.5 for every input of 10 floats. Flux has a function called train! to do all this for us.
data, labels = rand(10, 100), fill(0.5, 2, 100) #dataset
loss(x, y) = sum(Flux.crossentropy(m(x), y)) #creating loss function
Flux.train!(loss, params(m), [(data,labels)], opt) #training the model You don’t have to use the train! In cases where arbitrary logic might be better suited, you could open up this training loop like so:
for d in training_set #assuming d looks like ( data, labels)
# our logic here
gs = gradient( params( m ) ) do # m is our model
l = loss(d...)
end
update!( opt, params(m), gs)
endAnd this concludes the basics of Flux usage, in the next section, we will learn to implement it to train a classifier for the CIFAR10 dataset.
Training a Classifier for the Deep Learning Model
Getting a real classifier to work might help fix the workflow in Julia a bit more. CIFAR10 is a dataset of 50k tiny training images split into 10 classes of dogs, birds, deer, etc. The reader is requested to check the image below for more details.
We will do the following steps in order to get a classifier trained –
- Load the dataset of CIFAR10 (both training and test dataset)
- Create a Convolution Neural Network (CNN)
- Define a loss function to calculate losses
- Use training data to train our network
- Evaluate our model on the test dataset
Useful Libraries to install before we proceed, installation is simple but might take few minutes to completely install.
] add Metalhead #to get the data ] add Images #Image processing package ] add ImageIO #to output images
Loading the Dataset
Metalhead.jl (Package) is an excellent package that has tons of classic predefined and pre-trained CV (computer vision) models. It also consists of a variety of data loaders that come in handy during the dataset load process.
using Statistics
using Flux, Flux.Optimise #deep learning framework
using Metalhead, Images #to load dataset
using Metalhead: trainimgs
using Images.ImageCore #to work on image processing
using Flux: onehotbatch, onecold #to encode
using Base.Iterators: partition
using CUDA #for GPU functionalityThis image will give us an idea of the different types of labels we are dealing with.

Metalhead.download(CIFAR10) #download the dataset CIFAR10
X = trainimgs(CIFAR10) #take the training dataset as X
labels = onehotbatch([X[i].ground_truth.class for i in 1:50000],1:10) #encode the datasetTo get more information about what we are dealing with let’s take a look at a random image from the dataset.
image(x) = x.img # handy for use later
ground_truth(x) = x.ground_truth
image.(X[rand(1:end, 10)]) #to show the images in IJulia itself
With 3 RGB layers of the matrix (32x32x3), together create the image vector we see above. Now since the dataset is too large, we can pass them in batches (take 1000) and keep a set for validation to check the evaluation of our model. This process of passing them in batches is called mini-batch learning and is very popular in machine learning. So, in layman terms, rather than sending our entire dataset which is big and might not fit in RAM, we break the dataset into small packets (mini-batches), usually chosen randomly, and then train our model on it. It is observed that they help with escaping the saddle points (it is the minimax point on the surface of the curve).
First, we define a ‘getarray’ function that would help in converting the matrices to Float type.
getarray(X) = float.( permutedims( channelview( X ), (2, 3, 1))) #get the matrix to float type
imgs = [ getarray(X[i].img ) for i in 1:50000] #get all the matrices into floatIn our batch of 1000, the first 49,000 images will make our training set and the rest will be saved for validation or test set. To achieve this we can use the function called ‘partition’ which handily breaks down the set we give it in consecutive parts (1000). and to concatenate we use use ‘cat’ function along any dimension.
train = ( [ (cat(imgs[i]..., dims = 4), labels[:,i]) for i in partition(1:49000, 1000)]) |> gpu
valset = 49001:50000
valX = cat(imgs[valset]..., dims = 4) |> gpu
valY = labels[:, valset] |> gpuDefining the Classifier
Now comes the part where we can define our Convolutional Neural Network (CNN).
Definition of a convolutional neural network is – one that defines a kernel and slides it across a matrix to create an intermediate representation to extract features from. As it goes into deeper layers it creates higher-order features which make it suitable for images (although it can be used in plenty of other situations), where the structure of the subject is what will help us determine which class it belongs to.
m = Chain( #crearting a CNN Conv((5,5), 3=>16, relu), MaxPool((2,2)), #first layer of CNN Conv((5, 5), 16=>8, relu), MaxPool((2, 2)), #second layer of CNN x -> reshape(x, :, size(x, 4)), #reshaping data to feed it into Dense layers for classification Dense(200, 120), #first layer Dense(120, 84), #second layer Dense(84, 10), #third and final layer with 10 classification labels. softmax) |> gpu
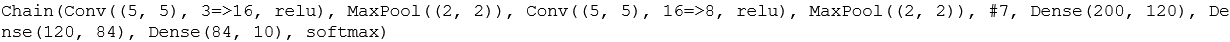
For the choice of the optimizer, we will be using momentum with cross-entropy.
Whenever we have to work with data that has multiple independent classes, cross-entropy comes in handy. And for the momentum, as the name suggests, it gradually lowers the learning rate as we proceed further with the training. This is necessary in case we overshoot from the desired destination and the chances for local minima increase while helping us to maintain a bit of adaptivity in our optimization.
using Flux: crossentropy, Momentum #import the optimizers
loss(x, y) = sum(crossentropy(m(x), y)) #using loss function
opt = Momentum(0.01) #fixing the momentum 
Before starting our training loop, we will need some sort of basic accuracy numbers about our model to keep the track of our progress. We can design our custom function to achieve just the same.
accuracy(x, y) = mean( onecold(m (x), 1:10) .== onecold(y, 1:10))

Training the Classifier
This is the part where we finally stitch everything together, here we do all the interesting operations which we defined previously to see what our model is capable of doing. Just for the tutorial, we will only be using 10 iterations over dataset (epochs) and optimize it, although for greater accuracy you can increase the epochs and play with hyperparameters a bit.
epochs = 10 #number of iterations
for epoch = 1:epochs
for d in train
gs = gradient(params(m)) do
l = loss(d...) #calculate losses
end
update!(opt, params(m), gs) #upadate the params weights
end
@show accuracy(valX, valY) #show the accuracy of model after each epoch
end
Step by step training process gives us a brief idea of how the network was learning the function. This accuracy is not bad at all for a model which was small and had no hyperparameter tuned with smaller epochs.
Training on a GPU
As you might have noticed we used GPU functions in our code previously as |> gpu to get (Flux) all these entities into the available GPU and get trained on it. The great thing about this is we just need that syntax and it will work on any piece of hardware. Just a small annotation and we get everything on GPU.
Testing the Network
As we have trained our neural network for 100 passes over the training dataset. But we would need to check if our model has learned anything at all. To check this, we simply predict the labels corresponding to each class from our neural net output, and checking it against the true values of class labels. If the prediction is correct, we add that sample to the correct prediction (true values) list. This will be done on the still unseen part of the data.
Firstly, we would have to get the same processing of images as we did on the training data set to compare them side by side.
valset = valimgs(CIFAR10) #value set
valimg = [ getarray(valset[i].img) for i in 1:10000] #get them to array
labels = onehotbatch([valset[i].ground_truth.class for i in 1:10000],1:10)#encode them
test = gpu.( [(cat(valimg[i]..., dims = 4), labels[:,i]) for i in partition(1:10000, 1000)])Next, we display some of the images from our validation dataset.
ids = rand(1:10000, 10) #random image ids
image.(valset[ids]) #show images in vector form
We have 10 values as the output for all 10 classes. If the particular value is higher for a class, our network thinks that image is from that particular class. The below image shows the values (energies) in 10 floats and every column corresponds to the output of one image.
Let’s see how our model fared on the dataset.
rand_test = getarray.( image.(valset[ids])) #get the test images
rand_test = cat(rand_test..., dims = 4) |> gpu # concat and feed to GPU
rand_truth = ground_truth.(valset[ids]) #check the values against true values
m(rand_test)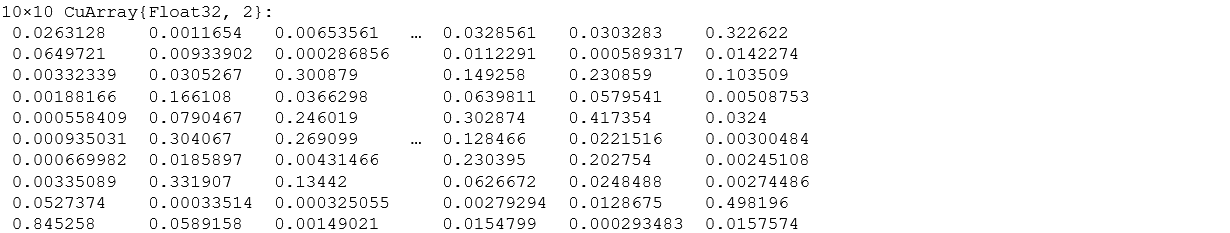
This looks very similar to how we would have expected our results to be. Even after the small training period, let’s see how our model actually performs on any new data given, (that was prepared by us).
accuracy( test[1]...)#testing accuracy
49% is clearly much better than the chances of randomly having it correct which is 10% (since we have 10 classes) which is not bad at all for the small hand-coded models without hyper-parameter tuning like ours.
Let’s take a look at how the net performed on all the classes performed individually.
class_correct = zeros(10) #creating an array of zeros
class_total = zeros(10)
for i in 1:10
preds = m(test[i][1]) #prediction after feeding it in our model
lab = test[i][2]
for j = 1:1000
pred_class = findmax(preds[:, j])[2] #find the argmax for each class
actual_class = findmax(lab[:, j])[2] #true vale of class
if pred_class == actual_class #if both are equal then then increment values by 1
class_correct[pred_class] += 1
end
class_total[actual_class] += 1
end
end
class_correct ./ class_total #getting total number of ratios (/100) times we get it correct
The spread seems pretty good, but some classes are performing significantly better than others. It is left for the reader to explore the reason.
Conclusion
In this article, we learned how powerful Julia is when it comes to computation. We learned about the Flux package and how to use it to train our hand-written model to classify between 10 different classes in just a few lines of code, that too on GPU!. We also learned about CuArrays and their significance in decreasing computation time. Hope this article has been helpful in starting your journey with Flux (Julia).
Thanks to the Mike Innes, Andrew Dinhobl, Ygor Canalli et al. for valuable documentation. Reach out to me via LinkedIn (Nihal Singh).

