Introduction
Artificial Intelligence (AI) has been making significant strides in various industries, and healthcare is no exception. One of the most promising areas within AI in healthcare is Natural Language Processing (NLP), which has the potential to revolutionize patient care by facilitating more efficient and accurate data analysis and communication.
NLP has proven to be a game changer in the field of healthcare. NLP is transforming the way healthcare providers deliver patient care. From population health management to disease detection, NLP is helping healthcare professionals make informed decisions and provide better treatment outcomes.
Learning Objectives
- Understanding and analyzing the use of NLP and AI in healthcare
- Getting a grip on the basics of NLP
- Getting to know about some commonly used NLP libraries in healthcare
- Learning about the use cases of NLP in healthcare
This article was published as a part of the Data Science Blogathon.
Table of Contents
- The Motivation for Using AI & NLP in Healthcare
- What is Natural Language Processing?
- Different Techniques Used in NLP
3.1 Rule-Based Techniques
3.2 Statistical Techniques using Machine Learning Models
3.3 Transfer Learning - Various NLP Libraries and Their Frameworks
- What are Large Language Models (LLM)?
- NLP in Clinical Text – The Need for Different Approach
- Some NLP Libraries Used in Healthcare Industry
- Understanding the Clinical Datasets
- What are Different Types of Clinical Data?
- Use Cases and Applications of NLP in the Healthcare Industry
- How to Build NLP Pipeline with Clinical Text?
11.1 Solution Design
11.2 Step-by-Step Code - Conclusion
The Motivation for Using AI & NLP in Healthcare
The motivation for using AI and NLP in healthcare is rooted in improving patient care and treatment outcomes while reducing healthcare costs. The healthcare industry generates vast amounts of data, including EMRs, clinical notes, and health-related social media posts, that can provide valuable insights into patient health and treatment outcomes. However, much of this data is unstructured and difficult to analyze manually.
Additionally, the healthcare industry faces several challenges, such as an aging population, increasing rates of chronic disease, and a shortage of healthcare professionals.
These challenges have led to a growing need for more efficient and effective healthcare delivery.
By providing valuable insights from unstructured medical data, NLP can help to improve patient care and treatment outcomes and support healthcare professionals in making more informed clinical decisions.
What is Natural Language Processing?
Natural Language Processing (NLP) is a subfield of Artificial Intelligence (AI) that deals with the interaction between computers and human languages. It uses computational techniques to analyze, understand, and generate human language.
Natural language processing is used in various applications, including speech recognition, machine translation, sentiment analysis, and text summarization. Text summarization using NLP automates the process of reducing lengthy text into shorter summaries, which is useful in domains such as financial research, media monitoring, and question-answer bots. This technique saves time and effort in comprehending complex texts while retaining essential information.
We will now explore the various NLP Techniques, libraries, and frameworks.
Different Techniques Used in NLP
There are two commonly used techniques used in the NLP industry.
1. Rule-based Techniques: rely on predefined grammar rules and dictionaries
2. Statistical Techniques: use machine learning algorithms to analyze and understand language
3. Large Language Model using Transfer Learning
Here is a standard NLP Pipeline with various NLP tasks
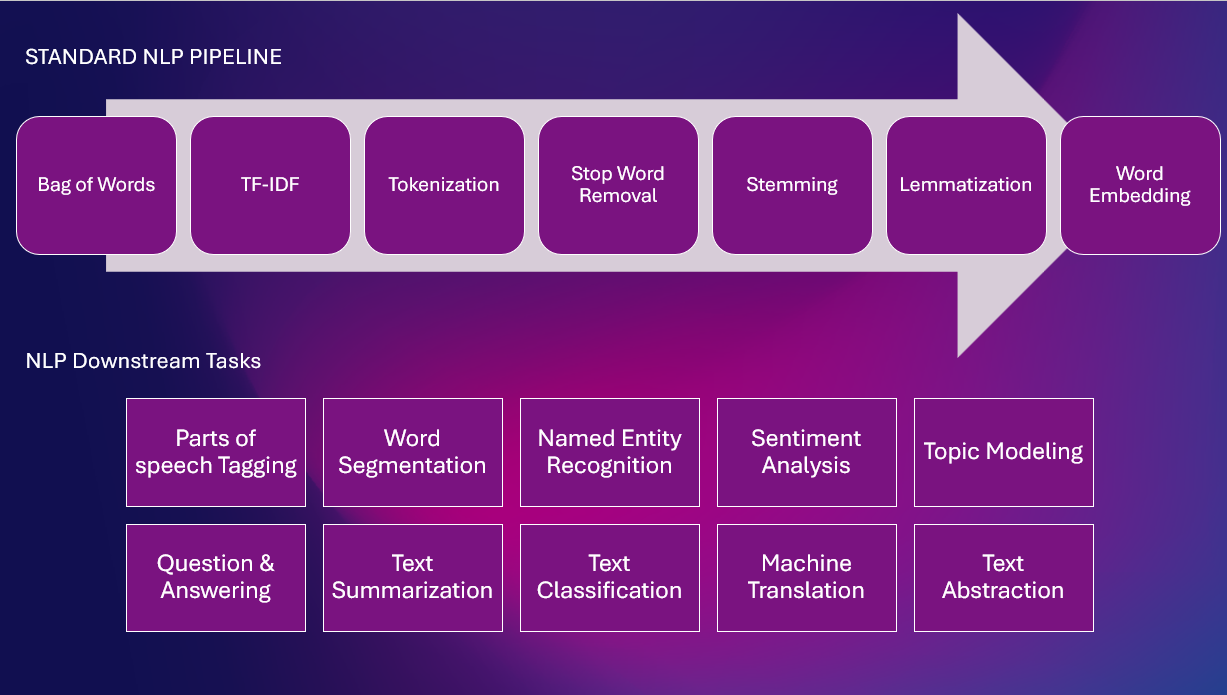
Rule-Based Techniques
These techniques involve creating a set of hand-crafted rules or patterns to extract meaningful information from text. Rule-based systems typically work by defining specific patterns that match the target information, such as named entities or specific keywords, and then extracting that information based on those patterns. Rule-based systems are fast, reliable, and straightforward, but they are limited by the quality and number of rules defined, and they can be difficult to maintain and update.
For example, a rule-based system for named entity recognition could be designed to identify proper nouns in text and categorize them into predefined entity types, such as a person, location, organization, disease, drugs, etc. The system would use a series of rules to identify patterns in the text that match the criteria for each entity type, such as capitalization for person names or specific keywords for organizations.
Statistical Techniques Using Machine Learning Models
These techniques use statistical algorithms to learn patterns in the data and make predictions based on those patterns. Machine learning models can be trained on large amounts of annotated data, making them more flexible and scalable than rule-based systems. Several types of machine learning models are used in NLP, including decision trees, random forests, support vector machines, and neural networks.
For example, a machine learning model for sentiment analysis could be trained on a large corpus of annotated text, where each text is tagged as positive, negative, or neutral. The model would learn the statistical patterns in the data that distinguish between positive and negative text and then use those patterns to make predictions on new, unseen text. The advantage of this approach is that the model can learn to identify sentiment patterns that are not explicitly defined in the rules.
Transfer Learning
These techniques are a hybrid approach combining the strengths of rule-based and machine-learning models. Transfer learning uses a pre-trained machine learning model, such as a language model trained on a large corpus of text, as a starting point for fine-tuning a specific task or domain. This approach leverages the general knowledge learned from the pre-trained model, reducing the amount of labeled data required for training and allowing for faster and more accurate predictions on a specific task.
For example, a transfer learning approach to named entity recognition could fine-tune a pre-trained language model on a smaller corpus of annotated medical text. The model would start with the general knowledge learned from the pre-trained model and then adjust its weights to match the medical text’s patterns better. This approach would reduce the amount of labeled data required for training and result in a more accurate model for named entity recognition in the medical domain.
Various NLP Libraries and Their Frameworks
Various libraries provide a wide range of NLP functionalities. Such as :
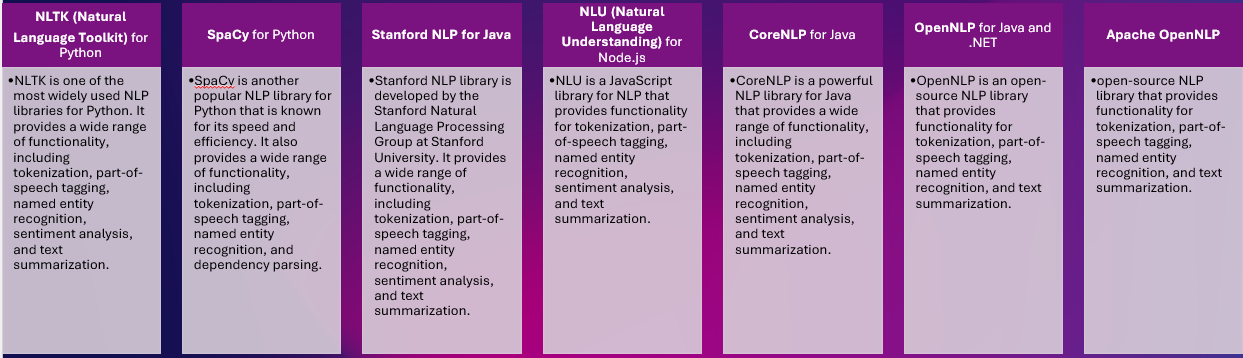
Natural Language Processing (NLP) libraries and frameworks are software tools that help develop and deploy NLP applications. Several NLP libraries and frameworks are available, each with strengths, weaknesses, and focus areas.
These tools vary in terms of the complexity of the algorithms they support, the size of the models they can handle, the ease of use, and the degree of customization they allow.
What are Large Language Models (LLM)?
Large language models are trained on massive amounts of data. Can generate human-like text and perform a wide range of NLP tasks with high accuracy.
Here are some examples of large language models and a brief description of each:
GPT-3 (Generative Pretrained Transformer 3): Developed by OpenAI, GPT-3 is a large transformer-based language model that uses deep learning algorithms to generate human-like text. It has been trained on a massive corpus of text data, allowing it to generate coherent and contextually appropriate text responses based on a prompt.
BERT (Bidirectional Encoder Representations from Transformers): Developed by Google, BERT is a transformer-based language model that has been pre-trained on a large corpus of text data. It is designed to perform well on a wide range of NLP tasks, such as named entity recognition, question answering, and text classification, by encoding the context and relationships between words in a sentence.
RoBERTa (Robustly Optimized BERT Approach): Developed by Facebook AI, RoBERTa is a variant of BERT that has been fine-tuned and optimized for NLP tasks. It has been trained on a larger corpus of text data and uses a different training strategy than BERT, leading to improved performance on NLP benchmarks.
ELMo (Embeddings from Language Models): Developed by Allen Institute for AI, ELMo is a deep contextualized word representation model that uses a bidirectional LSTM (Long Short-Term Memory) network to learn language representations from a large corpus of text data. ELMo can be fine-tuned for specific NLP tasks or used as a feature extractor for other machine-learning models.
ULMFiT (Universal Language Model Fine-Tuning): Developed by FastAI, ULMFiT is a transfer learning method that fine-tunes a pre-trained language model on a specific NLP task using a small amount of task-specific annotated data. ULMFiT has achieved state-of-the-art performance on a wide range of NLP benchmarks and is considered a leading example of transfer learning in NLP.
NLP in Clinical Text: The Need for Different Approach
Clinical text is often unstructured and contains a lot of medical jargon and acronyms, making it difficult for traditional NLP models to understand and process. Additionally, clinical text often includes important information such as disease, drugs, patient information, diagnoses, and treatment plans, which require specialized NLP models that can accurately extract and understand this medical information.
Another reason clinical text needs different NLP models is that it contains a large amount of data spread across different sources, such as EHRs, clinical notes, and radiology reports, which need to be integrated. This requires models that can process and understand the text and link and integrate the data across different sources and establish clinically acceptable relationships.
Lastly, clinical text often contains sensitive patient information and needs to be protected by strict regulations such as HIPAA. NLP models used to process clinical text must be able to identify and protect sensitive patient information while still providing useful insights.Given the sensitive nature of clinical text, including patient information, adhering to HIPAA regulations is Crucial. A HIPAA-compliant survey tool can help healthcare providers securely gather patient data while ensuring privacy.
Some NLP Libraries Used in Healthcare Industry
The textual data within medicine requires a specialized Natural Language Processing (NLP) system capable of extracting medical information from various sources such as clinical texts and other medical documents.
Here is a list of NLP libraries and models specific to the medical domain:
spaCy: It is an open-source NLP library that provides out-of-the-box models for various domains, including the medical domain.
ScispaCy: A specialized version of spaCy that is trained specifically on scientific and biomedical text, which makes it ideal for processing medical text.
BioBERT: A pre-trained transformer-based model specifically designed for the biomedical domain. It is pre-trained with Wiki + Books + PubMed + PMC.
ClinicalBERT: Another pre-trained model designed to process clinical notes & discharge summaries from the MIMIC-III database.
Med7: A transformer-based model that was trained on electronic health records (EHR) to extract seven key clinical concepts, including diagnosis, medication, and laboratory tests.
DisMod-ML: A probabilistic modeling framework for disease modeling that uses NLP techniques to process medical text.
MEDIC: A rule-based NLP system for extracting medical information from text.
These are some of the popular NLP libraries and models that are specifically designed for the medical domain. They offer a range of features, from pre-trained models to rule-based systems, and can help healthcare organizations process medical text effectively.
In our NER model, we will use spaCy and Scispacy. These libraries are comparatively easy to run on Google colab or local infrastructure.
The BioBERT and ClinicalBERT resource-intensive large language models need GPUs and higher infrastructure.
Understanding the Clinical Datasets
Medical text data can be obtained from various sources, such as electronic health records (EHRs), medical journals, clinical notes, medical websites, and databases. Some of these sources provide publicly available datasets that can be used for training NLP models, while others may require approval and ethical considerations before accessing the data. The sources of medical text data include:
1. Open-source medical corpora such as the MIMIC-III database is a large, openly accessible electronic health records (EHRs) database from patients who received care at the Beth Israel Deaconess Medical Center between 2001 and 2012. The database includes information such as patient demographics, vital signs, laboratory tests, medications, procedures, and notes from healthcare professionals, such as nurses and physicians. Additionally, the database includes information on patients’ ICU stays, including the type of ICU, length of stay, and outcomes. The data in MIMIC-III is de-identified and can be used for research purposes to support the development of predictive models and clinical decision support systems.
2. The National Library of Medicine’s ClinicalTrials.gov website has clinical trial data & disease surveillance data.
3. National Institutes of Health’s National Library of Medicine, National Centers for Biotechnology Information (NCBI), and the World Health Organization (WHO)
4. Healthcare institutions and organizations such as hospitals, clinics, and pharmaceutical companies generate large amounts of medical text data through electronic health records, clinical notes, medical transcription, and medical reports.
5. Medical research journals and databases, such as PubMed and CINAHL, contain vast amounts of published medical research articles and abstracts.
6. Social media platforms like Twitter can provide real-time insights into patient perspectives, drug reviews, and experiences.
To train NLP models using medical text data, it is important to consider the data’s quality and relevance and ensure that it is properly pre-processed and formatted. Additionally, it is important to adhere to ethical and legal considerations when working with sensitive medical information.
What are Different Types of Clinical Data?
Several types of clinical data are commonly used in healthcare:

Clinical data refers to information about individuals’ healthcare, including patient medical history, diagnoses, treatments, lab results, imaging studies, and other relevant health information.
EHR/EMR data are linked to Demographic data (This includes personal information such as age, gender, ethnicity, and contact information.), Patient-generated data (This type of data is generated by patients themselves, including information collected through patient-reported outcome measures and patient-generated health data.)
Other sets of data are :
Genomic Data: This type relates to an individual’s genetic information, including DNA sequences and markers.
Wearable Device Data: This data includes information collected from wearable devices such as fitness trackers and heart monitors.
Each type of clinical data plays a unique role in providing a comprehensive view of a patient’s health and is used in different ways by healthcare providers and researchers to improve patient care and inform treatment decisions.
Use Cases and Applications of NLP in the Healthcare Industry
Natural Language Processing (NLP) has been widely adopted in the healthcare industry and has several use cases. Some of the prominent ones include:
Population Health: NLP can be used to process large amounts of unstructured medical data such as medical records, surveys, and claims data to identify patterns, correlations, and insights. This helps in monitoring population health and early detection of diseases.
Patient Care: NLP can be used to process patients’ electronic health records (EHRs) to extract vital information such as diagnosis, medications, and symptoms. This information can be used to improve patient care and provide personalized treatment.
Disease Detection: NLP can be used to process large amounts of text data, such as scientific articles, news articles, and social media posts, to detect outbreaks of infectious diseases.
Clinical Decision Support System (CDSS): NLP can be used to analyze patients’ electronic health records to provide real-time decision support to healthcare providers. This helps in providing the best possible treatment options and improving the overall quality of care.
Clinical Trial: NLP can process clinical trial data to identify correlations and potential new treatments.
Drugs Adverse Events: NLP can be used to process large amounts of drug safety data to identify adverse events and drug interactions.
Precision Health: NLP can be used to process genomic data and medical records to identify personalized treatment options for individual patients.
Medical Professional’s Efficiency Improvement: NLP can automate routine tasks such as medical coding, data entry, and claim processing, freeing medical professionals to focus on providing better patient care.
These are just a few examples of how NLP revolutionizes the healthcare industry. As NLP technology continues to advance, we can expect to see more innovative uses of NLP in healthcare in the future.
How to Build NLP Pipeline with Clinical Text?
We will develop a step-by-step Spacy pipeline using SciSpacy NER Model for Clinical Text.
Objective: This project aims to construct an NLP pipeline utilizing SciSpacy to perform custom Named Entity Recognition on clinical texts.
Outcome: The outcome will be extracting information regarding diseases, drugs, and drug doses from clinical text, which can then be utilized in various NLP downstream applications.
Solution Design:
Here is the high-level solution to extract entity information from Clinical Text. NER extraction is important NLP task used in most of the NLP pipelines.
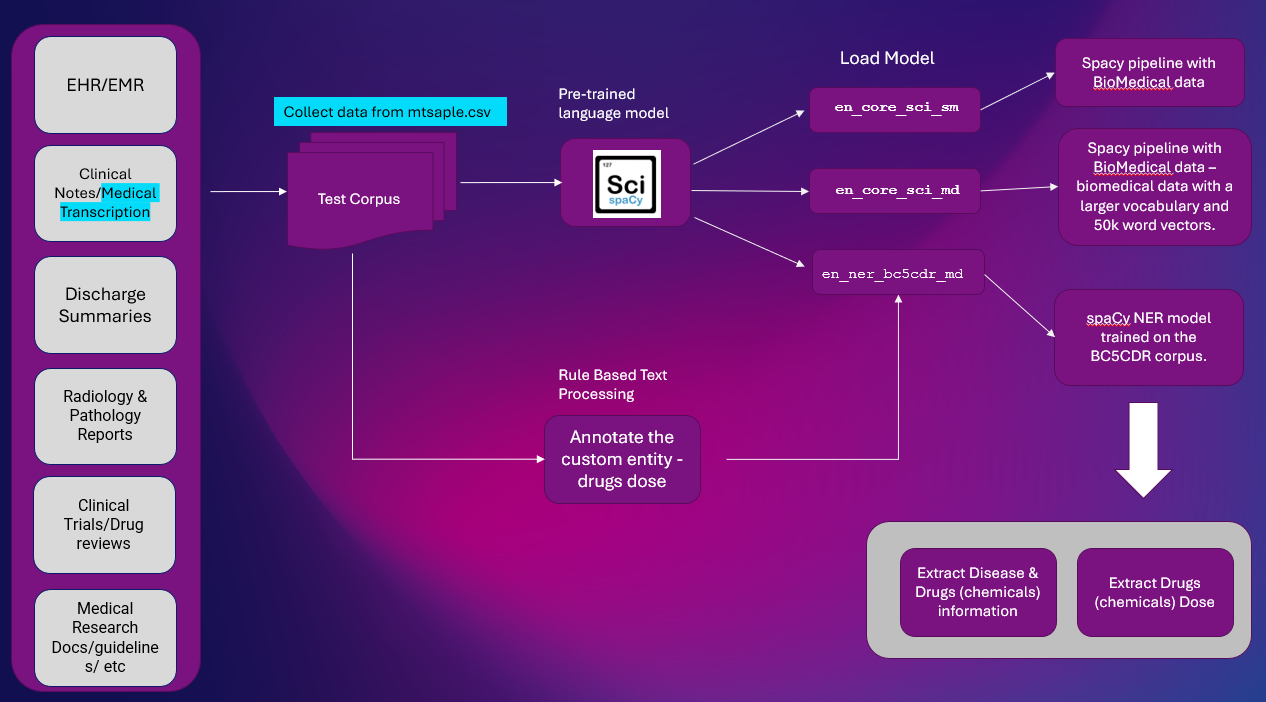
Platform: Google Colab
NLP Libraries: spaCy & SciSpacy
Dataset: mtsample.csv (scrapped data from mtsample).
We have used ScispaCy pre-trained NER model en_ner_bc5cdr_md-0.5.1 to extract disease and drugs. Drugs are extracted as Chemicals.
en_ner_bc5cdr_md-0.5.1 is a spaCy model for named entity recognition (NER) in the biomedical domain.
The “bc5cdr” refers to the BC5CDR corpus, a biomedical text corpus used to train the model. The “md” in the name refers to the biomedical domain. The “0.5.1” in the name refers to the version of the model.
We will use the sample “transcription” text from mtsample.csv and annotate using a rule-based pattern to extract drug doses.
Step-by-Step Code:
Install spacy & scispacy Packages. spaCy models are designed to perform specific NLP tasks, such as tokenization, part-of-speech tagging, and named entity recognition.
!pip install -U spacy !pip install scispacy
Install scispacy base models and NER models
The en_ner_bc5cdr_md-0.5.1 model is specifically designed to recognize named entities in biomedical text, such as diseases, genes, and drugs, as chemicals.
This model can be useful for NLP tasks in the biomedical domain, such as information extraction, text classification, and question-answering.
!pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.5.1/en_core_sci_sm-0.5.1.tar.gz !pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.5.1/en_core_sci_md-0.5.1.tar.gz !pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.5.1/en_ner_bc5cdr_md-0.5.1.tar.gz
Install other Packages
pip install render
Import Packages
import scispacy import spacy #Core models import en_core_sci_sm import en_core_sci_md
#NER specific models import en_ner_bc5cdr_md #Tools for extracting & displaying data from spacy import displacy import pandas as pd
Python Code:
import pandas as pd
mtsample_df=pd.read_csv("mtsamples.csv")
print(mtsample_df.shape)
print(mtsample_df.info())
print(mtsample_df.head(2))Test the models with sample data
# Pick specific transcription to use (row 3, column "transcription") and test the scispacy NER model text = mtsample_df.loc[10, "transcription"]

Load specific model: en_core_sci_sm and pass text through
nlp_sm = en_core_sci_sm.load() doc = nlp_sm(text)
#Display resulting
entity extraction displacy_image = displacy.render(doc, jupyter=True,style='ent')

Note the entity is tagged here. Mostly medicalterms. However, these are generic entities.
nlp_md = en_core_sci_md.load() doc = nlp_md(text)
#Display resulting entity extraction
displacy_image = displacy.render(doc, jupyter=True,style='ent')
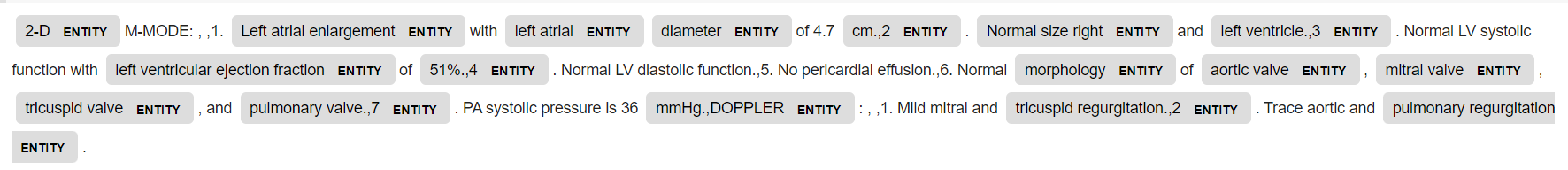
This time the numbers are also tagged as entities by en_core_sci_md.
Now Load specific model: import en_ner_bc5cdr_md and pass text through
nlp_bc = en_ner_bc5cdr_md.load() doc = nlp_bc(text) #Display resulting entity extraction displacy_image = displacy.render(doc, jupyter=True,style='ent')

Now two medical entities are tagged: disease and chemical(drugs).
Display the entity
print("TEXT", "START", "END", "ENTITY TYPE")
for ent in doc.ents:
print(ent.text, ent.start_char, ent.end_char, ent.label_)
TEXT START END ENTITY TYPE
Morbid obesity 26 40 DISEASE
Morbid obesity 70 84 DISEASE
weight loss 400 411 DISEASE
Marcaine 1256 1264 CHEMICAL
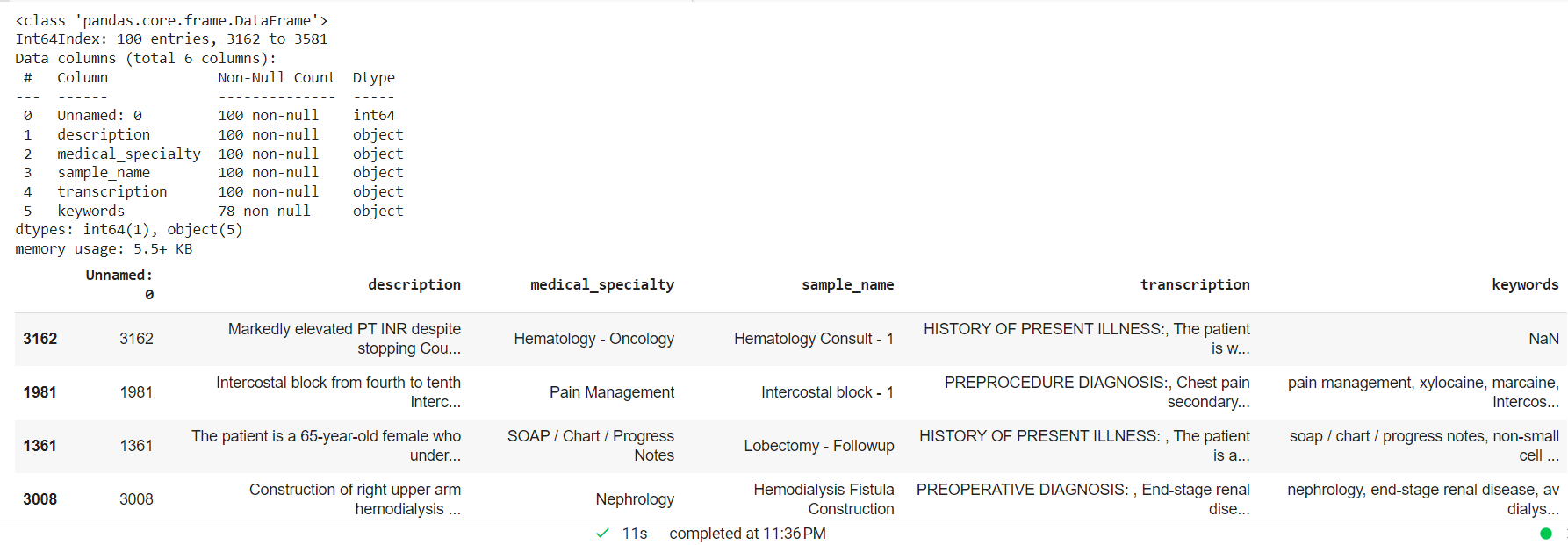
Process the clinical text dropping NAN values and creating a random smaller sample for the custom entity model.
mtsample_df.dropna(subset=['transcription'], inplace=True) mtsample_df_subset = mtsample_df.sample(n=100, replace=False, random_state=42) mtsample_df_subset.info() mtsample_df_subset.head()
spaCy matcher – The rule-based matching resembles the usage of regular expressions, but spaCy provides additional capabilities. Using the tokens and relationships within a document enables you to identify patterns that include entities with the help of NER models. The goal is to locate drug names and their dosages from the text, which could help detect medication errors by comparing them with standards and guidelines.
The goal is to locate drug names and their dosages from the text, which could help detect medication errors by comparing them with standards and guidelines.
from spacy.matcher import Matcher
pattern = [{'ENT_TYPE':'CHEMICAL'}, {'LIKE_NUM': True}, {'IS_ASCII': True}]
matcher = Matcher(nlp_bc.vocab)
matcher.add("DRUG_DOSE", [pattern])
for transcription in mtsample_df_subset['transcription']:
doc = nlp_bc(transcription)
matches = matcher(doc)
for match_id, start, end in matches:
string_id = nlp_bc.vocab.strings[match_id] # get string representation
span = doc[start:end] # the matched span adding drugs doses
print(span.text, start, end, string_id,)
#Add disease and drugs
for ent in doc.ents:
print(ent.text, ent.start_char, ent.end_char, ent.label_)
The output will display the entities extracted from the clinical text sample.
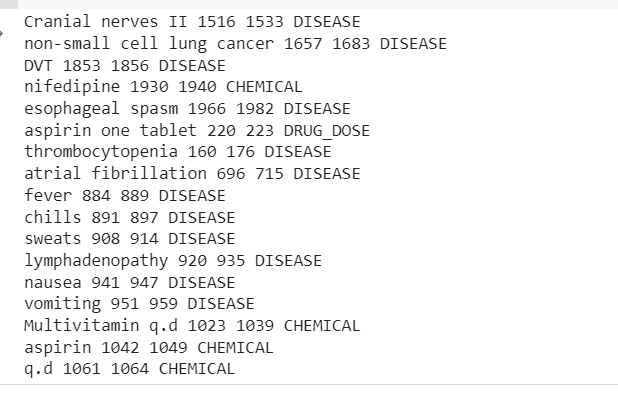
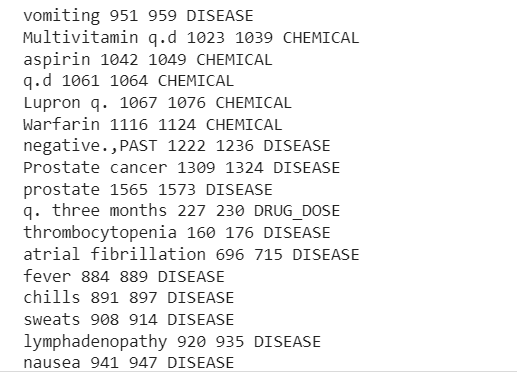
Now we can see the pipeline extracted Disease, Drugs(Chemicals), and Drugs-Doses information from the clinical text.
There is some misclassification, but we can increase the model’s performance using more data.
We can now use these medical entities in various tasks like disease detection, predictive analysis, clinical decision support system, medical text classification, summarization, questions -answering, and many more.
Conclusion
1. In this article, we have explored some of the key features of NLP in Healthcare, which will help to understand the complex healthcare text data.
We also implemented scispaCy and spaCy and constructed a simple custom NER model through a pre-trained NER model and rule-based matcher. While we have only covered one NER model, numerous others are available, and a vast amount of additional functionality to discover.
2. Within the scispaCy framework, there are numerous additional techniques to explore, including methods for detecting abbreviations, performing dependency parsing, and identifying individual sentences.
3. The latest trends in NLP for healthcare include the development of domain-specific models like BioBERT and ClinicalBert and using large language models like GPT-3. These models offer a high level of accuracy and efficiency, but their use also raises concerns about bias, privacy, and control over data.
ChatGPT (an advanced conversational AI model developed by OpenAI) is already making a huge impact in the NLP world. The model is trained on a massive amount of text data from the internet and has the capability to generate human-like text responses based on the input it receives. It can be used for various tasks such as question answering, summarization, translation, and more. The model is also fine-tuned for specific use cases, such as generating code or writing articles, to enhance its performance in those specific areas.
5. However, despite its numerous benefits, NLP in healthcare is not without its challenges. Ensuring the accuracy and fairness of NLP models and overcoming data privacy concerns are some of the challenges that need to be addressed to fully realize the potential of NLP in healthcare.
6. With its many advantages, it is essential for healthcare professionals to embrace and incorporate NLP into their workflows. While there are many challenges to overcome, NLP in healthcare is certainly a trend worth watching and investing in.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.







