Introduction
Retrieval Augmented Generation has been here for a while. Many tools and applications are being built around this concept, like vector stores, retrieval frameworks, and LLMs, making it convenient to work with custom documents, especially Semi-structured Data with Langchain. Working with long, dense texts has never been so easy and fun. The conventional RAG works well with unstructured text-heavy files like DOC, PDFs, etc. However, this approach does not sit well with semi-structured data, such as embedded tables in PDFs.
While working with semi-structured data, there are usually two concerns.
- The conventional extraction and text-splitting methods do not account for tables in PDFs. They usually end up breaking up the tables. Hence resulting in information loss.
- Embedding tables may not translate to precise semantic search.
So, in this article, we will build a Retrieval generation pipeline for semi-structured data with Langchain to address these two concerns with semistructured data.
Learning Objectives
- Understand the difference between structured, unstructured, and semi-structured data.
- A mild refresher on Retrieval Augement Generation and Langchain.
- Learn how to build a multi-vector retriever to handle semi-structured data with Langchain.
This article was published as a part of the Data Science Blogathon.
Table of contents
Types of Data
There are usually three types of data. Structured, Semi-structured, and Unstructured.
- Structured Data: The structured data is the standardized data. The data follows a pre-defined schema, such as rows and columns. SQL databases, Spreadsheets, data frames, etc.
- Unstructured Data: Unstructured data, unlike structured data, follows no data model. The data is as random as it can get. For example, PDFs, Texts, Images, etc.
- Semi-structured Data: It is the combination of the former data types. Unlike the structured data, it does not have a rigid pre-defined schema. However, the data still maintains a hierarchical order based on some markers, which is in contrast to unstructured types. For example, CSVs, HTML, Embedded tables in PDFs, XMLs, etc.

What is RAG?
RAG stands for Retrieval Augmented Generation. It is the simplest way to feed the Large language models with novel information. So, let’s have a quick primer on RAG.
In a typical RAG pipeline, we have knowledge sources, such as local files, Web pages, databases, etc, an embedding model, a vector database, and an LLM. We collect the data from various sources, split the documents, get the embeddings of text chunks, and store them in a vector database. Now, we pass the embeddings of queries to the vector store, retrieve the documents from the vector store, and finally generate answers with the LLM.
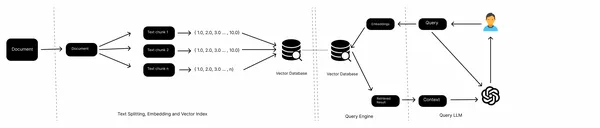
This is a workflow of a conventional RAG and works well with unstructured data like texts. However, when it comes to semi-structured data, for example, embedded tables in a PDF, it often fails to perform well. In this article, we will learn how to handle these embedded tables.
What is Langchain?
The Langchain is an open-source framework for building LLM-based applications. Since its launch, the project has garnered wide adoption among software developers. It provides a unified range of tools and technologies to build AI applications faster. Langchain houses tools such as vector stores, document loaders, retrievers, embedding models, text splitters, etc. It is a one-stop solution for building AI applications. But there is two core value proposition that makes it stand apart.
- LLM chains: Langchain provides multiple chains. These chains chain together several tools to accomplish a single task. For example, ConversationalRetrievalChain chains together an LLM, Vector store retriever, embedding model, and a chat history object to generate responses for a query. The tools are hard coded and have to be defined explicitly.
- LLM agents: Unlike LLM chains, AI agents do not have hard-coded tools. Instead of chaining one tool after another, we let the LLM decide which one to select and when based on text descriptions of tools. This makes it ideal for building complex LLM applications involving reasoning and decision-making.
Building The RAG pipeline
Now that we have a primer on the concepts. Let’s discuss the approach to building the pipeline. Working with semi-structured data can be tricky as it does not follow a conventional schema for storing information. And to work with unstructured data, we need specialized tools tailor-made for extracting information. So, in this project, we will use one such tool called “unstructured”; it is an open-source tool for extracting information from different unstructured data formats, such as tables in PDFs, HTML, XML, etc. Unstructured uses Tesseract and Poppler under the hood to process multiple data formats in files. So, let’s set up our environment and install dependencies before diving into the coding part.
Set-up Dev Env
Like any other Python project, open a Python environment and install Poppler and Tesseract.
!sudo apt install tesseract-ocr
!sudo apt-get install poppler-utilsNow, install the dependencies that we will need in our project.
!pip install "unstructured[all-docs]" Langchain openaiExtract Using Unstructured
Now that we have installed the dependencies, we will extract data from a PDF file.
from unstructured.partition.pdf import partition_pdf
pdf_elements = partition_pdf(
"mistral7b.pdf",
chunking_strategy="by_title",
extract_images_in_pdf=True,
max_characters=3000,
new_after_n_chars=2800,
combine_text_under_n_chars=2000,
image_output_dir_path="./"
)Running it will install several dependencies like YOLOx that are needed for OCR and return object types based on extracted data. Enabling extract_images_in_pdf will let unstructured extract embedded images from files. This can help implement multi-modal solutions.
Now, let’s explore the categories of elements from our PDF.
# Create a dictionary to store counts of each type
category_counts = {}
for element in pdf_elements:
category = str(type(element))
if category in category_counts:
category_counts[category] += 1
else:
category_counts[category] = 1
# Unique_categories will have unique elements
unique_categories = set(category_counts.keys())
category_countsRunning this will output element categories and their count.
Now, we separate the elements for easy handling. We create an Element type that inherits from Langchain’s Document type. This is to ensure more organized data, which is easier to deal with.
from unstructured.documents.elements import CompositeElement, Table
from langchain.schema import Document
class Element(Document):
type: str
# Categorize by type
categorized_elements = []
for element in pdf_elements:
if isinstance(element, Table):
categorized_elements.append(Element(type="table", page_content=str(element)))
elif isinstance(element, CompositeElement):
categorized_elements.append(Element(type="text", page_content=str(element)))
# Tables
table_elements = [e for e in categorized_elements if e.type == "table"]
# Text
text_elements = [e for e in categorized_elements if e.type == "text"]Multi-vector Retriever
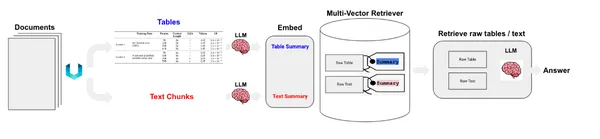
We have table and text elements. Now, there are two ways we can handle these. We can store the raw elements in a document store or store summaries of texts. Tables might pose a challenge to semantic search; in that case, we create the summaries of tables and store them in a document store along with the raw tables. To achieve this, we will use MultiVectorRetriever. This retriever will manage a vector store where we store the embeddings of summary texts and a simple in-memory document store to store raw documents.
First, build a summarizing chain to summarize the table and text data we extracted earlier.
from langchain.chat_models import cohere
from langchain.prompts import ChatPromptTemplate
from langchain.schema.output_parser import StrOutputParser
prompt_text = """You are an assistant tasked with summarizing tables and text. \
Give a concise summary of the table or text. Table or text chunk: {element} """
prompt = ChatPromptTemplate.from_template(prompt_text)
model = cohere.ChatCohere(cohere_api_key="your_key")
summarize_chain = {"element": lambda x: x} | prompt | model | StrOutputParser()
tables = [i.page_content for i in table_elements]
table_summaries = summarize_chain.batch(tables, {"max_concurrency": 5})
texts = [i.page_content for i in text_elements]
text_summaries = summarize_chain.batch(texts, {"max_concurrency": 5})I have used Cohere LLM for summarizing data; you may use OpenAI models like GPT-4. Better models will yield better outcomes. Sometimes, the models may not perfectly capture table details. So, it is better to use capable models.
Now, we create the MultivectorRetriever.
from langchain.retrievers import MultiVectorRetriever
from langchain.prompts import ChatPromptTemplate
import uuid
from langchain.embeddings import OpenAIEmbeddings
from langchain.schema.document import Document
from langchain.storage import InMemoryStore
from langchain.vectorstores import Chroma
# The vectorstore to use to index the child chunks
vectorstore = Chroma(collection_name="collection",
embedding_function=OpenAIEmbeddings(openai_api_key="api_key"))
# The storage layer for the parent documents
store = InMemoryStore()
id_key = ""id"
# The retriever
retriever = MultiVectorRetriever(
vectorstore=vectorstore,
docstore=store,
id_key=id_key,
)
# Add texts
doc_ids = [str(uuid.uuid4()) for _ in texts]
summary_texts = [
Document(page_content=s, metadata={id_key: doc_ids[i]})
for i, s in enumerate(text_summaries)
]
retriever.vectorstore.add_documents(summary_texts)
retriever.docstore.mset(list(zip(doc_ids, texts)))
# Add tables
table_ids = [str(uuid.uuid4()) for _ in tables]
summary_tables = [
Document(page_content=s, metadata={id_key: table_ids[i]})
for i, s in enumerate(table_summaries)
]
retriever.vectorstore.add_documents(summary_tables)
retriever.docstore.mset(list(zip(table_ids, tables)))
We used Chroma vector store for storing summary embeddings of texts and tables and an in-memory document store to store raw data.
RAG
Now that our retriever is ready, we can build an RAG pipeline using Langchain Expression Language.
from langchain.schema.runnable import RunnablePassthrough
# Prompt template
template = """Answer the question based only on the following context,
which can include text and tables::
{context}
Question: {question}
"""
prompt = ChatPromptTemplate.from_template(template)
# LLM
model = ChatOpenAI(temperature=0.0, openai_api_key="api_key")
# RAG pipeline
chain = (
{"context": retriever, "question": RunnablePassthrough()}
| prompt
| model
| StrOutputParser()
)
Now, we can ask questions and receive answers based on retrieved embeddings from the vector store.
chain.invoke(input = "What is the MT bench score of Llama 2 and Mistral 7B Instruct??")Conclusion
A lot of information stays hidden in semi-structured data format. And it is challenging to extract and perform conventional RAG on these data. In this article, we went from extracting texts and embedded tables in the PDF to building a multi-vector retriever and RAG pipeline with Langchain. So, here are the key takeaways from the article.
Key Takeaways
- Conventional RAG often faces challenges dealing with semi-structured data, such as breaking up tables during text splitting and imprecise semantic searches.
- Unstructured, an open-source tool for semi-structured data, can extract embedded tables from PDFs or similar semi-structured data.
- With Langchain, we can build a multi-vector retriever for storing tables, texts, and summaries in document stores for better semantic search.
Frequently Asked Questions
A: Semi-structured data, unlike structured data, does not have a rigid schema but has other forms of markers to enforce hierarchies.
A. Semi-structured data examples are CSV, Emails, HTML, XML, parquet files, etc.
A. LangChain is an open-source framework that simplifies the creation of applications using large language models. It can be used for various tasks, including chatbots, RAG, question-answering, and generative tasks.
A. A RAG pipeline retrieves documents from external data stores, processes them to store them in a knowledge base, and provides tools to query them.
A. Llama Index explicitly designs search and retrieval applications, while Langchain offers flexibility for creating custom AI agents.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.





