Introduction
While working with Pandas in Python, you may often notice that your code is slow, it uses too much memory, or it gets difficult to handle as your data grows larger. Any of these issues can lead to errors, long wait times for analysis, and limitations in processing bigger datasets. In this article, we will explore different ways to make computation happen faster for the code you have written in Pandas.
The tricks mentioned in this guide will mature your project code and save you a lot of time. The computational time you save will reflect or translate into cost savings, better user experience, and better operational experience. This is beneficial when deploying your ML or Python code into a cloud environment such as Amazon web services (AWS), Google Cloud Platform (GCP), or Microsoft Azure.

Table of Contents
Making Pandas Faster Using Numba
This first method is Numba which speeds up your code that uses Numpy underneath. It is an open-source JIT(Just in Time) compiler. You can use Numba for mathematical operations, NumPy codes, and multiple loops. We are specifying Numpy codes because Pandas are built on top of NumPy. Let’s see how to use Numba:
Get Personalized Learning Path! Set your goal and timeline. Get a path—under 2 mins.
1. Install Numba
!pip install numba ==0.53.12. Import Libraries
import numba
from numba import jit
print(numba.version)3. Define Task
Get the list of prime numbers from 0 to 100
Let’s code without using Numba
# Define function to extract prime numbers from a given list using simple Python
def check_prime(y):
prime_numbers = []
for num in y:
flag = False
if num > 1:
# check for factors
for i in range(2, num):
if (num % i) == 0:
# if factor is found, set flag to True
flag = True
# break out of loop
break
if flag == False:
prime_numbers.append(num)
return prime_numbers4. Calculate Time
Let’s calculate the time taken to perform the task
# List of 0 to 100
x = np.arange(100)
x
# DO NOT REPORT THIS... COMPILATION TIME IS INCLUDED IN THE EXECUTION TIME!
start = time.time()
check_prime(x)
end = time.time()
print("Elapsed (with compilation) = %s" % (end - start))
# NOW THE FUNCTION IS COMPILED, RE-TIME IT EXECUTING FROM CACHE
start = time.time()
check_prime(x)
end = time.time()
print("Elapsed (after compilation) = %s" % (end - start))Output

In a notebook, the %timeit magic function is the best to use because it runs the function many times in a loop to get a more accurate estimate of the execution time of short functions.
%timeit check_prime(x)Now, let’s code using Numba’s JIT decorator
# Define function to extract prime numbers from a given list using jit decorator and nopython = True mode
@jit(nopython=True)
def check_prime(y):
prime_numbers = []
for num in y:
flag = False
if num > 1:
# check for factors
for i in range(2, num):
if (num % i) == 0:
# if factor is found, set flag to True
flag = True
# break out of loop
break
if flag == False:
prime_numbers.append(num)
return np.array(prime_numbers)Let’s calculate the time taken to perform the task
# DO NOT REPORT THIS... COMPILATION TIME IS INCLUDED IN THE EXECUTION TIME!
start = time.time()
check_prime(x)
end = time.time()
print("Elapsed (with compilation) = %s" % (end - start))
# NOW THE FUNCTION IS COMPILED, RE-TIME IT EXECUTING FROM CACHE
start = time.time()
check_prime(x)
end = time.time()
print("Elapsed (after compilation) = %s" % (end - start))
In a notebook, the %timeit magic function is the best to use because it runs the function many times in a loop to get a more accurate estimate of the execution time of short functions.
%timeit check_prime(x)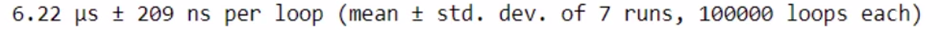
You can see that computation is very high using Numba.
Using Dask for Making Pandas Computation Faster
Dask provides efficient parallelization for data analytics in Python. Dask Dataframes allows you to work with large datasets for both data manipulation and building ML models with only minimal code changes. It is open source and works well with Python libraries like NumPy, sci-kit-learn, etc.
Why Dask?
Pandas is not sufficient when the data gets bigger, bigger than what you can fit in the RAM. You may use Spark or Hadoop to solve this. But, these are not Python environments. This stops you from using NumPy, sklearn, Pandas, TensorFlow, and all the commonly used Python libraries for ML. It scales up to clusters. This is where Dask comes to the rescue.
!pip install dask==2021.05.0
!pip install graphviz==0.16
!pip install python-graphviz
# Import dask
import dask
print(dask.__version__)Parallel Processing with Dask
Task
– Apply a discount of 20% to 2 products worth 100 and 200 respectively and generate a total bill
Function without Dask
Let’s define the functions. Since the task is very small, I am adding up sleep time of 1 second in every function.
from time import sleep
# Define functions to apply discount, get the total of 2 products, get the final price of 2 products
def apply_discount(x):
sleep(1)
x = x - 0.2 * x
return x
def get_total_price(a, b):
sleep(1)
return a + bLet’s calculate the total bill and note down the time taken for the task. I am using %%time function to note the time
%%time
product1 = apply_discount(100)
product2 = apply_discount(200)
total_bill = get_total_price(product1, product2)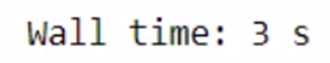
The total time taken for the above task is 4.01s. Let’s use Dask and check the time taken
Function with Dask
Use the delayed function from Dask to reduce the time
# Import dask.delayed
from dask import delayed
%%time
# Wrapping the function calls using dask.delayed
product1 = delayed(apply_discount)(100) # no work has happened yet
product2 = delayed(apply_discount)(200) # no work has happened yet
total_bill = delayed(get_total_price)(product1, product2) # no work has happened yet
total_bill
Delayed('get_total_price-d7ade4e9-d9ba-4a9f-886a-ec66b20c6d66')As you can see the total time taken with a delayed wrapper is only 374 µs. But the work hasn’t happened yet. A delayed wrapper creates a delayed object, that keeps track of all the functions to call and the arguments to pass to it. Basically, it has built a task graph that explains the entire computation. You don’t have the output yet.
Most Dask workloads are lazy, that is, they don’t start any work until you explicitly trigger them with a call to compute().
So let’s use compute() to get the output
total_bill.compute()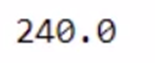
Now you have the output. This operation also took some time. Let’s compute the total time taken.
%%time
# Wrapping the function calls using dask.delayed
product1 = delayed(apply_discount)(100)
product2 = delayed(apply_discount)(200)
total_bill = delayed(get_total_price)(product1, product2)
total_bill.compute()
The total time taken is 2.01 seconds. It’s 1 second less than the original functions. Any idea how dask.delayed did this?
You can see the optimal task graph created by Dask by calling the visualize() function. Let’s see.
# Visualize the total_bill object
total_bill.visualize()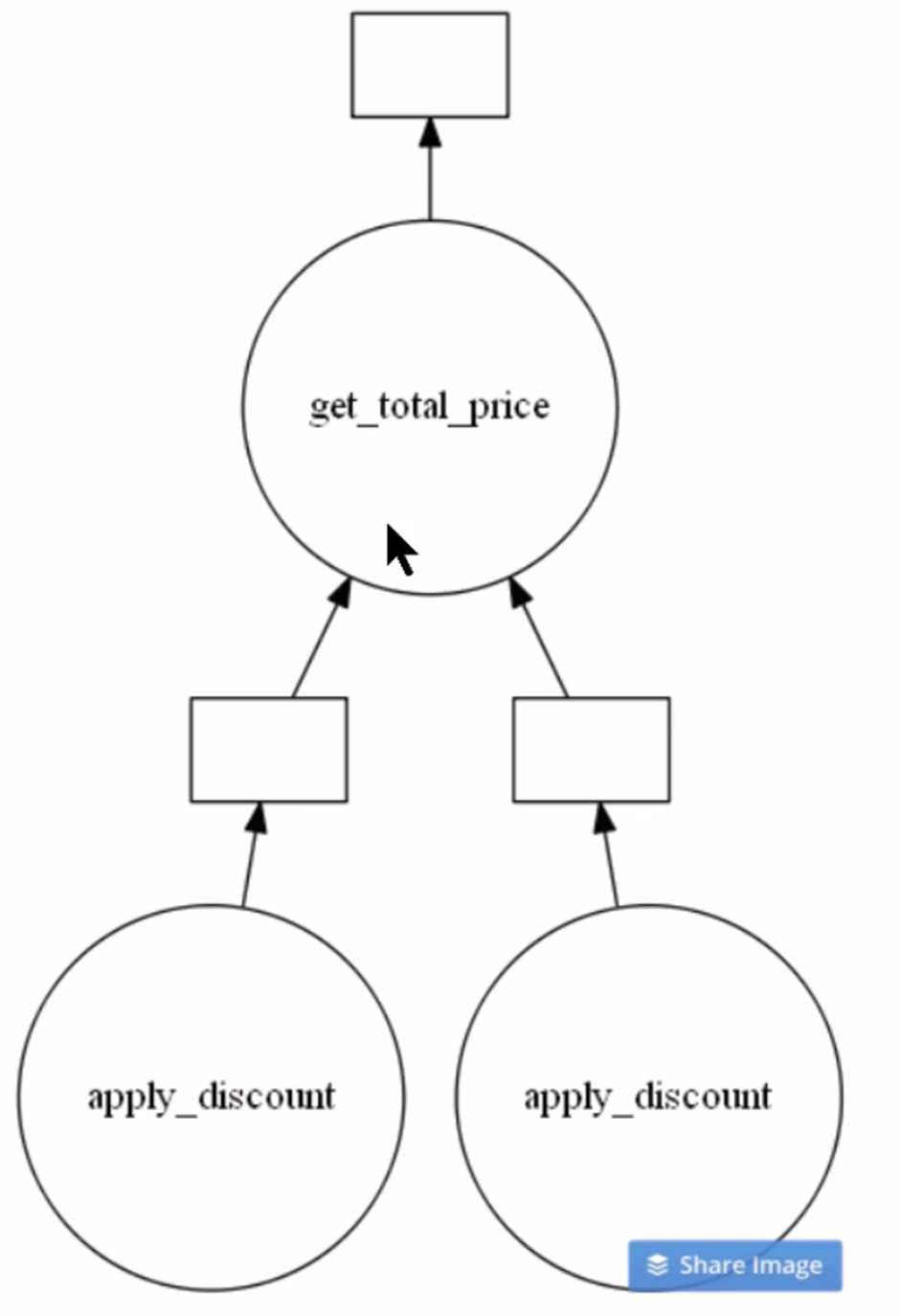
Clearly from the above image, you can see there are two instances of apply_discount() function called in parallel. This is an opportunity to save time and processing power by executing them simultaneously.
This was one of the most basic use cases of Dask.
Making Pandas Faster Using Modin
Modin is a Python library that can be used to handle large datasets using parallelization. It uses Ray or Dask to provide an effortless way to speed up operations.
The syntax is similar to Pandas and its astounding performance has made it a promising solution. All you have to do is change just one line of code.
# Install Modin dependencies and Dask to run on Dask
!pip install -U pandas
!pip install modin[dask]
!pip install "dask[distributed]"Modin also allows you to choose which engine you wish to use for computation. The environment variable MODIN_ENGINE is used for this. The below code shows how to specify the computation engine:
import os
os.environ["MODIN_ENGINE"] = "ray" # Modin will use Ray
os.environ["MODIN_ENGINE"] = "dask" # Modin will use DaskBy default, Modin will use all of the cores available on your system.
But if you wish to limit the cores as you wish to do some other task, you can limit the number of CPUs Modin uses with the below command:
import os
os.environ["MODIN_CPUS"] = "4"
import modin.pandas as pdTask
- Load the dataset using Pandas and Modin and compare the total time taken.
I am going to use pandas_pd for pandas and modin_pd for modin. First, let’s load the data using Pandas.
# Load Pandas and time
import pandas as pandas_pd
import time
# Load csv file using pandas
%time pandas_df = pandas_pd.read_csv("your_dataset.csv")
Initialize Dask client
# For Dask backend
from dask.distributed import Client
client = Client()
# Load csv file using modin pd
%time modin_df = pd.read_csv("Datasets/large_dataset.csv")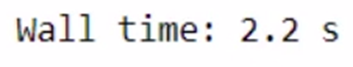
The total time taken is 1 min 10 seconds. The time taken might vary as per the specification of your system. We were able to save a few seconds only for such a small task. Imagine how much time we can save while working on a bigger dataset and a lot of computations!
Let’s perform a few tasks using Modin.
Task
- Print the head of both dataframes and compare the time
# pandas df fillna
%time pandas_df.fillna(0)
# modin df fillna
%time modin_df.fillna(0)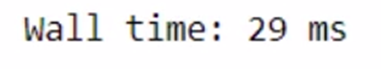
Modin is taking lesser time.
Conclusion
In this article, we have seen how different Python functions are used to speed up the computation speed of Pandas code. We learned how Numba is used for optimized numerical computations, while Dask is used in parallel processing on massive datasets. We also saw how Modin’s familiar Pandas-like interface speeds up through Ray or Dask. Choosing the best approach amongst these depends on your data size, computation type, and desired level of ease-use.
Stay tuned for another article where we share more tips and tricks to make your Python Pandas code faster and more efficient!






