Hey there, fellow Python enthusiast! Have you ever wished your NumPy code run at supersonic speed? Meet JAX!. Your new best friend in your machine learning, deep learning, and numerical computing journey. Think of it as NumPy with superpowers. It can automatically handle gradients, compile your code to run fast using JIT, and even run on GPU and TPU without breaking a sweat. Whether you’re building neural networks, crunching scientific data, tweaking transformer models, or just trying to speed up your calculations, JAX has your back. Let’s dive in and see what makes JAX so special.
This guide provides a detailed introduction to JAX and its ecosystem.
Learning Objectives
- Explain JAX’s core principles and how they differ from Numpy.
- Apply JAX’s three key transformations to optimize Python code. Convert NumPy operations into efficient JAX implementation.
- Identify and fix common performance bottlenecks in JAX code. Implement JIT compilation correctly while avoiding typical Pitfalls.
- Build and train a Neural Network from scratch using JAX. Implement common machine learning operations using JAX’s functional approach.
- Solve optimization problems using JAX’s automatic differentiation. Perform efficient matrix operations and numerical computations.
- Apply effective debugging strategies for JAX-specific issues. Implement memory-efficient patterns for large-scale computations.
This article was published as a part of the Data Science Blogathon.
Table of contents
What is JAX?
According to the official documentation, JAX is a Python library for acceleration-oriented array computation and program transformation, designed for high-performance numerical computing and large-scale machine learning. So, JAX is essentially NumPy on steroids, It combines familiar NumPy-style operations with automatic differentiation and hardware acceleration. Think of it as getting the best of three worlds.
- NumPy’s elegant syntax and array operation
- PyTorch like automatic differentiation capability
- XLA’s (Accelerated Linear Algebra) for hardware acceleration and compilation benefits.
Why does JAX Stand Out?
What sets JAX apart is its transformations. These are powerful functions that can modify your Python code:
- JIT: Just-In-Time compilation for faster execution
- Grad: Automatic differentiation for computing gradients
- vmap: Automatically vectorization for batch processing
Here is a quick look:
import jax.numpy as jnp
from jax import grad, jit
# Define a simple function
@jit # Speed it up with compilation
def square_sum(x):
return jnp.sum(jnp.square(x))
# Get its gradient function automatically
gradient_fn = grad(square_sum)
# Try it out
x = jnp.array([1.0, 2.0, 3.0])
print(f"Gradient: {gradient_fn(x)}")Output:
Gradient: [2. 4. 6.]Getting Started with JAX
Below we will follow some steps to get started with JAX.
Step1: Installation
Setting up JAX is straightforward for CPU-only use. You can use the JAX documentation for more information.
Step2: Creating Environment for Project
Create a conda environment for your project
# Create a conda env for jax
$ conda create --name jaxdev python=3.11
#activate the env
$ conda activate jaxdev
# create a project dir name jax101
$ mkdir jax101
# Go into the dir
$cd jax101
Step3: Installing JAX
Installing JAX in the newly created environment
# For CPU only
pip install --upgrade pip
pip install --upgrade "jax"
# for GPU
pip install --upgrade pip
pip install --upgrade "jax[cuda12]"
Now you are ready to dive into real things. Before getting your hands dirty on practical coding let’s learn some new concepts. I will be explaining the concepts first and then we will code together to understand the practical viewpoint.
First, get some motivation, By the way, why do we learn a new library again? I will answer that question throughout this guide in a step-by-step manner as simple as possible.
Why Learn JAX?
Think of JAX as a power tool. While NumPy is like a reliable hand saw, JAX is like a modern electric saw. It requires a bit more steps and knowledge, but the performance benefits are worth it for intensive computation tasks.
- Performance: Jax code can run significantly faster than Pure Python or NumPy code, especially on GPU and TPUs
- Flexibility: It’s not just for machine learning- JAX excels in scientific computing, optimization, and simulation.
- Modern Approach: JAX encourages functional programming patterns that lead to cleaner, more maintainable code.
In the next section, we’ll dive deep into JAX’s transformation, starting with the JIT compilation. These transformations are what give JAX its superpowers, and understanding them is key to leveraging JAX effectively.
Essential JAX Transformations
JAX’s transformations are what truly set it apart from the numerical computation libraries such as NumPy or SciPy. Let’s explore each one and see how they can supercharge your code.
JIT or Just-In-Time Compilation
Just-in-time compilation optimizes code execution by compiling parts of a program at runtime rather than ahead of time.
How JIT works in JAX?
In JAX, jax.jit transforms a Python function into a JIT-compiled version. Decorating a function with @jax.jit captures its execution graph, optimizes it, and compiles it using XLA. The compiled version then executes, delivering significant speedups, especially for repeated function calls.
Here is how you can try it.
import jax.numpy as jnp
from jax import jit
import time
# A computationally intensive function
def slow_function(x):
for _ in range(1000):
x = jnp.sin(x) + jnp.cos(x)
return x
# The same function with JIT
@jit
def fast_function(x):
for _ in range(1000):
x = jnp.sin(x) + jnp.cos(x)
return x
Here is the same function, one is just a plain Python compilation process and the other one is used as a JAX’s JIT compilation process. It will calculate the 1000 data points sum of sine and cosine functions. we will compare the performance using time.
# Compare performance
x = jnp.arange(1000)
# Warm-up JIT
fast_function(x) # First call compiles the function
# Time comparison
start = time.time()
slow_result = slow_function(x)
print(f"Without JIT: {time.time() - start:.4f} seconds")
start = time.time()
fast_result = fast_function(x)
print(f"With JIT: {time.time() - start:.4f} seconds")
The result will astonish you. The JIT compilation is 333 times faster than the normal compilation. It’s like comparing a bicycle with a Buggati Chiron.
Output:
Without JIT: 0.0330 seconds
With JIT: 0.0010 secondsJIT can give you a superfast execution boost but you must use it properly otherwise it will be like driving Bugatti on a muddy village road that offers no supercar facility.
Common JIT Pitfalls
JIT works best with static shapes and types. Avoid using Python loops and conditions that depend on array values. JIT does not work with the dynamic arrays.
# Bad - uses Python control flow
@jit
def bad_function(x):
if x[0] > 0: # This won't work well with JIT
return x
return -x
# print(bad_function(jnp.array([1, 2, 3])))
# Good - uses JAX control flow
@jit
def good_function(x):
return jnp.where(x[0] > 0, x, -x) # JAX-native condition
print(good_function(jnp.array([1, 2, 3])))
Output:

That means bad_function is bad because JIT was not located in the value of x during calculation.
Output:
[1 2 3]Limitations and Considerations
- Compilation Overhead: The first time a JIT-compiled function is executed, there is some overhead due to compilation. The compilation cost may outweigh the performance benefits for small functions or those called only once.
- Dynamic Python Features: JAX’s JIT requires functions to be “static”. Dynamic control flow, like changing shapes or values based on Python loops, is not supported in the compiled code. JAX provided alternatives like `jax.lax.cond` and `jax.lax.scan` to handle dynamic control flow.
Automatic Differentiation
Automatic differentiation, or autodiff, is a computation technique for calculating the derivative of functions accurately and effectively. It plays a crucial role in optimizing machine learning models, especially in training neural networks, where gradients are used to update model parameters.

How does Automatic differentiation work in JAX?
Autodiff works by applying the chain rule of calculus to decompose complex functions into simpler ones, calculating the derivative of these sub-functions, and then combining the results. It records each operation during the function execution to construct a computational graph, which is then used to compute derivatives automatically.
There are two main modes of auto-diff:
- Forward Mode: Computes derivatives in a single forward pass through the computational graph, efficient for functions with a small number of parameters.
- Reverse Mode: Computes derivatives in a single backward pass through the computational graph, efficient for functions with a large number of parameters.
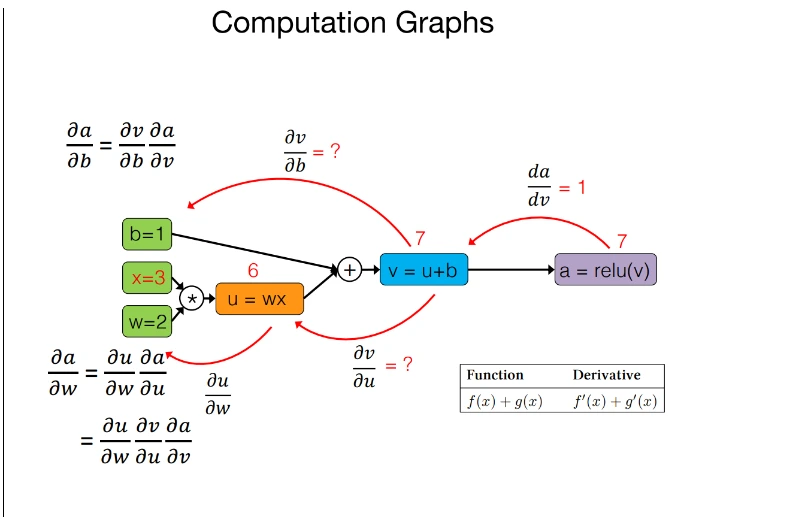
Key features in JAX automatic differentiation
- Gradient Computation(jax.grad): `jax.grad` computes the derivative of a scaler-output function for its input. For functions with multiple inputs, a partial derivative can be obtained.
- Higher-Order Derivative(jax.jacobian, jax.hessian) : JAX supports the computation of higher-order derivatives, such as Jacobians and Hessains, making it suitable for advanced optimization and physics simulation.
- Composability with other JAX Transformation: Autodiff in JAX integrates seamlessly with other transformations like `jax.jit` and `jax.vmap` allowing for efficient and scalable computation.
- Reverse-Mode Differentiation(Backpropagation): JAX’s auto-diff uses reverse-mode differentiation for scaler-output functions, which is highly effective for deep learning tasks.
import jax.numpy as jnp
from jax import grad, value_and_grad
# Define a simple neural network layer
def layer(params, x):
weight, bias = params
return jnp.dot(x, weight) + bias
# Define a scalar-valued loss function
def loss_fn(params, x):
output = layer(params, x)
return jnp.sum(output) # Reducing to a scalar
# Get both the output and gradient
layer_grad = grad(loss_fn, argnums=0) # Gradient with respect to params
layer_value_and_grad = value_and_grad(loss_fn, argnums=0) # Both value and gradient
# Example usage
key = jax.random.PRNGKey(0)
x = jax.random.normal(key, (3, 4))
weight = jax.random.normal(key, (4, 2))
bias = jax.random.normal(key, (2,))
# Compute gradients
grads = layer_grad((weight, bias), x)
output, grads = layer_value_and_grad((weight, bias), x)
# Multiple derivatives are easy
twice_grad = grad(grad(jnp.sin))
x = jnp.array(2.0)
print(f"Second derivative of sin at x=2: {twice_grad(x)}")
Output:
Second derivatives of sin at x=2: -0.9092974066734314Effectiveness in JAX
- Efficiency: JAX’s automatic differentiation is highly efficient due to its integration with XLA, allowing for optimization at the machine code level.
- Composability: The ability to combine different transformations makes JAX a powerful tool for building complex machine learning pipelines and Neural Networks architecture such as CNN, RNN, and Transformers.
- Ease of Use: JAX’s syntax for autodiff is simple and intuitive, enabling users to compute gradient without delving into the details of XLA and complex library APIs.
JAX Vectorize Mapping
In JAX, `vmap` is a powerful function that automatically vectorizes computations, allowing you to apply a function over batches of data without manually writing loops. It maps a function over an array axis (or multiple axes) and evaluates it efficiently in parallel, which can lead to significant performance improvements.
How vmap Works in JAX?
The vmap function automates the process of applying a function to each element along a specified axis of an input array while preserving the efficiency of the computation. It transforms the given function to accept batched inputs and execute the computation in a vectorized manner.
Instead of using explicit loops, vmap allows operations to be performed in parallel by vectorizing over an input axis. This leverages the hardware’s capability to perform SIMD (Single Instruction, Multiple Data) operations, which can result in substantial speed-ups.
Key Features of vmap
- Automatic Vectorization: vamp automates the batching of computations, making it simple to parallel code over batch dimensions without changing the original function logic.
- Composability with other Transformations: It works seamlessly with other JAX transformations, such as jax.grad for differentiation and jax.jit for Just-In-Time compilation, allowing for highly optimized and flexible code.
- Handling Multiple Batch Dimensions: vmap supports mapping over multiple input arrays or axes, making it versatile for various use cases like processing multi-dimensional data or multiple variables simultaneously.
import jax.numpy as jnp
from jax import vmap
# A function that works on single inputs
def single_input_fn(x):
return jnp.sin(x) + jnp.cos(x)
# Vectorize it to work on batches
batch_fn = vmap(single_input_fn)
# Compare performance
x = jnp.arange(1000)
# Without vmap (using a list comprehension)
result1 = jnp.array([single_input_fn(xi) for xi in x])
# With vmap
result2 = batch_fn(x) # Much faster!
# Vectorizing multiple arguments
def two_input_fn(x, y):
return x * jnp.sin(y)
# Vectorize over both inputs
vectorized_fn = vmap(two_input_fn, in_axes=(0, 0))
# Or vectorize over just the first input
partially_vectorized_fn = vmap(two_input_fn, in_axes=(0, None))
# print
print(result1.shape)
print(result2.shape)
print(partially_vectorized_fn(x, y).shape)
Output:
(1000,)
(1000,)
(1000,3)Effectiveness of vmap in JAX
- Performance Improvements: By vectorizing computations, vmap can significantly speed up execution by leveraging parallel processing capabilities of modern hardware like GPUs, and TPUs(Tensor processing units).
- Cleaner Code: It allows for more concise and readable code by eliminating the need for manual loops.
- Compatibility with JAX and Autodiff: vmap can be combined with automatic differentiation (jax.grad), allowing for the efficient computation of derivatives over batches of data.
When to Use Each Transformation
Using @jit when:
- Your function is called multiple times with similar input shapes.
- The function contains heavy numerical computations.
Use grad when:
- You need derivatives for optimization.
- Implementing machine learning algorithms
- Solving differential equations for simulations
Use vmap when:
- Processing batches of data with.
- Parallelizing computations
- Avoiding explicit loops
Matrix Operations and Linear Algebra Using JAX
JAX provides comprehensive support for matrix operations and linear algebra, making it suitable for scientific computing, machine learning, and numerical optimization tasks. JAX’s linear algebra capabilities are similar to those found in libraries like NumPY but with additional features such as automatic differentiation and Just-In-Time compilation for optimized performance.
Matrix Addition and Subtraction
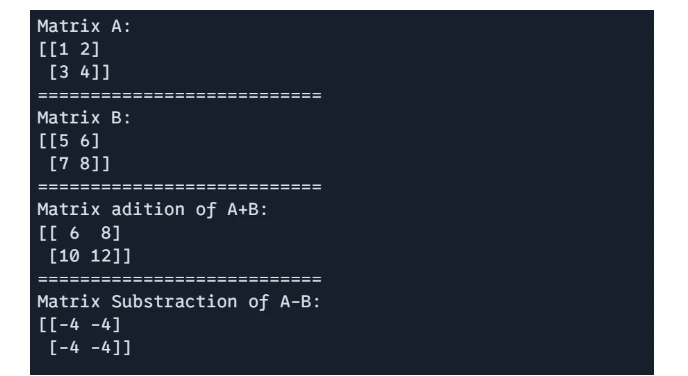
These operation are performed element-wise matrices of the same shape.
# 1 Matrix Addition and Subtraction:
import jax.numpy as jnp
A = jnp.array([[1, 2], [3, 4]])
B = jnp.array([[5, 6], [7, 8]])
# Matrix addition
C = A + B
# Matrix subtraction
D = A - B
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix B: \n{B}")
print("===========================")
print(f"Matrix adition of A+B: \n{C}")
print("===========================")
print(f"Matrix Substraction of A-B: \n{D}")
Output:
Matrix Multiplication
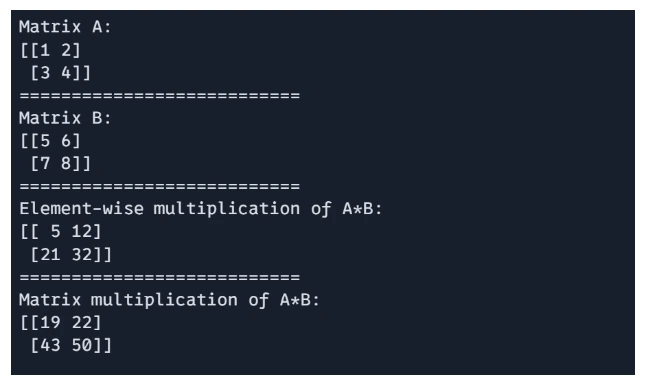
JAX support both element-wise multiplication and dor product-based matrix multiplication.
# Element-wise multiplication
E = A * B
# Matrix multiplication (dot product)
F = jnp.dot(A, B)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix B: \n{B}")
print("===========================")
print(f"Element-wise multiplication of A*B: \n{E}")
print("===========================")
print(f"Matrix multiplication of A*B: \n{F}")
Output:
Matrix Transpose
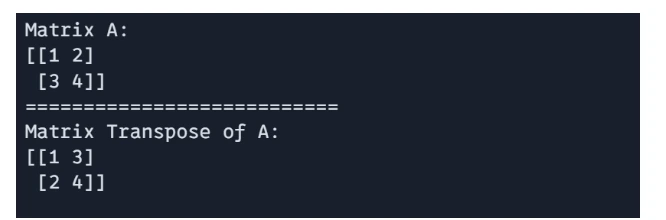
The transpose of a matrix can be obtained using `jnp.transpose()`
# Matric Transpose
G = jnp.transpose(A)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix Transpose of A: \n{G}")
Output:
Matrix Inverse
JAX provides function for matrix inversion using `jnp.linalg.inv()`
# Matric Inversion
H = jnp.linalg.inv(A)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix Inversion of A: \n{H}")
Output:
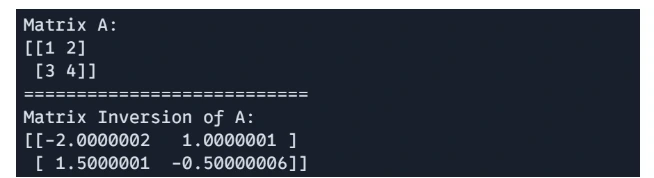
Matrix Determinant
Determinant of a matrix can be calculate using `jnp.linalg.det()`.
# matrix determinant
det_A = jnp.linalg.det(A)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix Determinant of A: \n{det_A}")
Output:
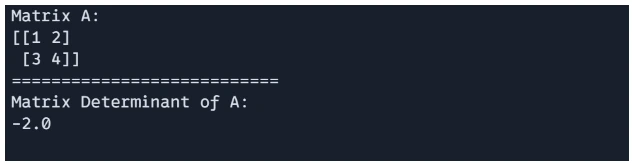
Matrix Eigenvalues and Eigenvectors
You can compute the eigenvalues and eigenvectors of a matrix using `jnp.linalg.eigh()`
# Eigenvalues and Eigenvectors
import jax.numpy as jnp
A = jnp.array([[1, 2], [3, 4]])
eigenvalues, eigenvectors = jnp.linalg.eigh(A)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Eigenvalues of A: \n{eigenvalues}")
print("===========================")
print(f"Eigenvectors of A: \n{eigenvectors}")
Output:
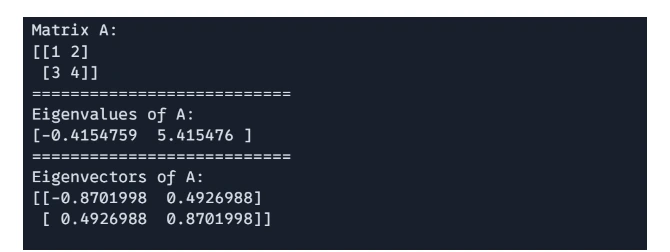
Matrix Singular Value Decomposition
SVD is supported via `jnp.linalg.svd`, useful in dimensionality reduction and matrix factorization.
# Singular Value Decomposition(SVD)
import jax.numpy as jnp
A = jnp.array([[1, 2], [3, 4]])
U, S, V = jnp.linalg.svd(A)
print(f"Matrix A: \n{A}")
print("===========================")
print(f"Matrix U: \n{U}")
print("===========================")
print(f"Matrix S: \n{S}")
print("===========================")
print(f"Matrix V: \n{V}")
Output:
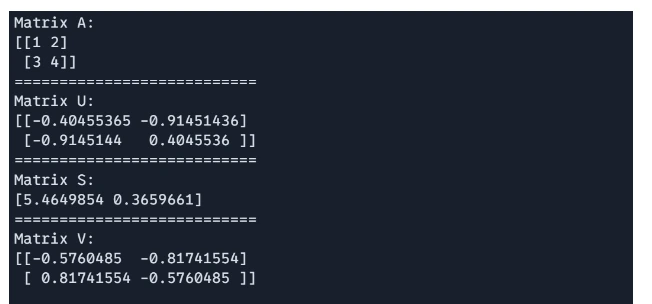
Solving System of Linear Equations
To solve a system of linear equation Ax = b, we use `jnp.linalg.solve()`, where A is a square matrix and b is a vector or matrix of the same number of rows.
# Solving system of linear equations
import jax.numpy as jnp
A = jnp.array([[2.0, 1.0], [1.0, 3.0]])
b = jnp.array([5.0, 6.0])
x = jnp.linalg.solve(A, b)
print(f"Value of x: {x}")Output:
Value of x: [1.8 1.4]Computing the Gradient of a Matrix Function
Using JAX’s automatic differentiation, you can compute the gradient of a scalar function with respect to a matrix.
We will calculate gradient of the below function and values of X
Function
# Computing the Gradient of a Matrix Function
import jax
import jax.numpy as jnp
def matrix_function(x):
return jnp.sum(jnp.sin(x) + x**2)
# Compute the grad of the function
grad_f = jax.grad(matrix_function)
X = jnp.array([[1.0, 2.0], [3.0, 4.0]])
gradient = grad_f(X)
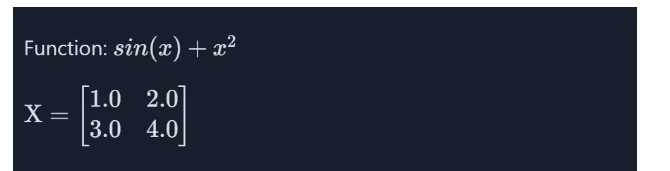
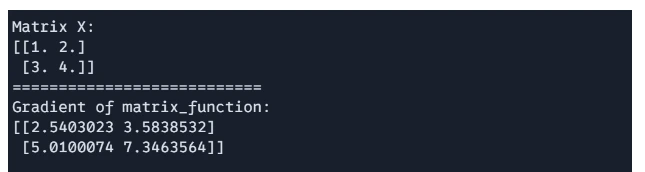
print(f"Matrix X: \n{X}")
print("===========================")
print(f"Gradient of matrix_function: \n{gradient}")
Output:
These most useful function of JAX used in numerical computing, machine learning, and physics calculation. There are many more left for you to explore.
Scientific Computing with JAX
JAX’s powerful libraries for scientific computing, JAX is best for scientific computing for its advance features such as JIT compilation, automatic differentiation, vectorization, parallelization, and GPU-TPU acceleration. JAX’s ability to support high performance computing makes it suitable for a wide range of scientific applications, including physics simulations, machine learning, optimization and numerical analysis.
We will explore an Optimization Problem in this section.
Optimization Problems
Let us go through the optimization problems steps below:
Step1: Define the function to minimize(or the problem)
# Define a function to minimize (e.g., Rosenbrock function)
@jit
def rosenbrock(x):
return sum(100.0 * (x[1:] - x[:-1] ** 2.0) ** 2.0 + (1 - x[:-1]) ** 2.0)Here, the Rosenbrock function is defined, which is a common test problem in optimization. The function takes an array x as input and computes a valie that represents how far x is from the function’s global minimum. The @jit decorator is used to enable Jut-In-Time compilation, which speed up the computation by compiling the function to run efficiently on CPUs and GPUs.
Step2: Gradient Descent Step Implementation
# Gradient descent optimization
@jit
def gradient_descent_step(x, learning_rate):
return x - learning_rate * grad(rosenbrock)(x)This function performs a single step of the gradient descent optimization. The gradient of the Rosenbrock function is calculated using grad(rosenbrock)(x), which provides the derivative with respects to x. The new value of x is updated by subtraction the gradient scaled by a learning_rate.The @jit is doing the same as before.
Step3: Running the Optimization Loop
# Optimize
x = jnp.array([0.0, 0.0]) # Starting point
learning_rate = 0.001
for i in range(2000):
x = gradient_descent_step(x, learning_rate)
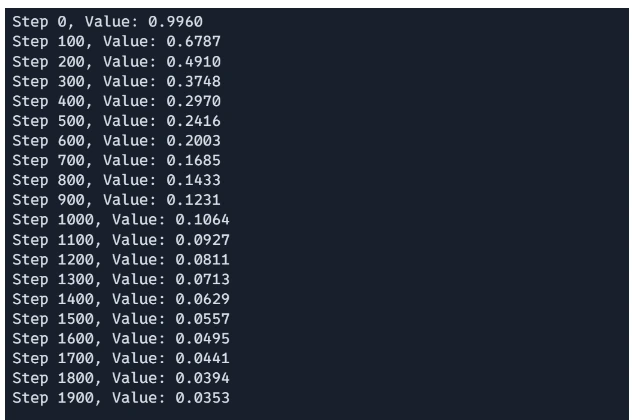
if i % 100 == 0:
print(f"Step {i}, Value: {rosenbrock(x):.4f}")The optimization loop initializes the starting point x and performs 1000 iterations of gradient descent. In each iteration, the gradient_descent_step function updates based on the current gradient. Every 100 steps, the current step number and the value of the Rosenbrock function at x are printed, providing the progress of the optimization.
Output:
Solving Real-world physics problem with JAX
We will simulate a physical system the motion of a damped harmonic oscillator, which models things like a mass-spring system with friction, shock absorbers in vehicles, or oscillation in electrical circuits. Is it not nice? Let’s do it.
Step1: Parameters Definition
import jax
import jax.numpy as jnp
# Define parameters
mass = 1.0 # Mass of the object (kg)
damping = 0.1 # Damping coefficient (kg/s)
spring_constant = 1.0 # Spring constant (N/m)
# Define time step and total time
dt = 0.01 # Time step (s)
num_steps = 3000 # Number of steps
The mass, damping coefficient, and spring constant are defined. These determine the physical properties of the damped harmonic oscillator.
Step2: ODE Definition
# Define the system of ODEs
def damped_harmonic_oscillator(state, t):
"""Compute the derivatives for a damped harmonic oscillator.
state: array containing position and velocity [x, v]
t: time (not used in this autonomous system)
"""
x, v = state
dxdt = v
dvdt = -damping / mass * v - spring_constant / mass * x
return jnp.array([dxdt, dvdt])The damped harmonic oscillator function defines the derivatives of the position and velocity of the oscillator, representing the dynamical system.
Step3: Euler’s Method
# Solve the ODE using Euler's method
def euler_step(state, t, dt):
"""Perform one step of Euler's method."""
derivatives = damped_harmonic_oscillator(state, t)
return state + derivatives * dt
A simple numerical method is used to solve the ODE. It approximates the state at the next time step on the basis of the current state and derivative.
Step4: Time Evolution Loops
# Initial state: [position, velocity]
initial_state = jnp.array([1.0, 0.0]) # Start with the mass at x=1, v=0
# Time evolution
states = [initial_state]
time = 0.0
for step in range(num_steps):
next_state = euler_step(states[-1], time, dt)
states.append(next_state)
time += dt
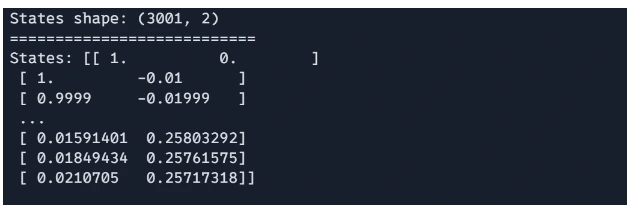
# Convert the list of states to a JAX array for analysis
states = jnp.stack(states)
The loop iterates through the specified number of time steps, updating the state at each step using Euler’s method.
Output:
Step5: Plotting The Results
Finally, we can plot the results to visualize the behavior of the damped harmonic oscillator.
# Plotting the results
import matplotlib.pyplot as plt
plt.style.use("ggplot")
positions = states[:, 0]
velocities = states[:, 1]
time_points = jnp.arange(0, (num_steps + 1) * dt, dt)
plt.figure(figsize=(12, 6))
plt.subplot(2, 1, 1)
plt.plot(time_points, positions, label="Position")
plt.xlabel("Time (s)")
plt.ylabel("Position (m)")
plt.legend()
plt.subplot(2, 1, 2)
plt.plot(time_points, velocities, label="Velocity", color="orange")
plt.xlabel("Time (s)")
plt.ylabel("Velocity (m/s)")
plt.legend()
plt.tight_layout()
plt.show()
Output:
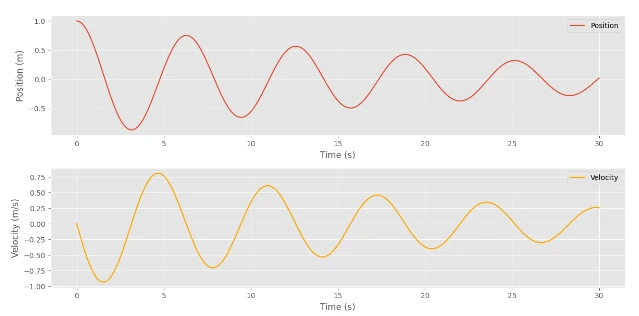
I know you are eager to see how the Neural Network can be built with JAX. So, let’s dive deep into it.
Here, you can see that the Values were minimized gradually.
Building Neural Networks with JAX
JAX is a powerful library that combines high-performance numerical computing with the ease of using NumPy-like syntax. This section will guide you through the process of constructing a neural network using JAX, leveraging its advanced features for automatic differentiation and just-in-time compilation to optimize performance.
Step1: Importing Libraries
Before we dive into building our neural network, we need to import the necessary libraries. JAX provides a suite of tools for creating efficient numerical computations, while additional libraries will assist with optimization and visualization of our results.
import jax
import jax.numpy as jnp
from jax import grad, jit
from jax.random import PRNGKey, normal
import optax # JAX's optimization library
import matplotlib.pyplot as pltStep2: Creating the Model Layers
Creating effective model layers is crucial in defining the architecture of our neural network. In this step, we’ll initialize the parameters for our dense layers, ensuring that our model starts with well-defined weights and biases for effective learning.
def init_layer_params(key, n_in, n_out):
"""Initialize parameters for a single dense layer"""
key_w, key_b = jax.random.split(key)
# He initialization
w = normal(key_w, (n_in, n_out)) * jnp.sqrt(2.0 / n_in)
b = normal(key_b, (n_out,)) * 0.1
return (w, b)
def relu(x):
"""ReLU activation function"""
return jnp.maximum(0, x)
- Initializing Function: init_layer_params initializes weights(w) and biases (b) for dense layers using He initialization for weight and a small value for biases. He or Kaiming He initialization works better for layers with ReLu activation functions, there are other popular initialization methods such as Xavier initialization which works better for layers with sigmoid activation.
- Activation Function: The relu function applies the ReLu activation function to the inputs which set negative values to zero.
Step3: Defining the Forward Pass
The forward pass is the cornerstone of a neural network, as it dictates how input data flows through the network to produce an output. Here, we will define a method to compute the output of our model by applying transformations to the input data through the initialized layers.
def forward(params, x):
"""Forward pass for a two-layer neural network"""
(w1, b1), (w2, b2) = params
# First layer
h1 = relu(jnp.dot(x, w1) + b1)
# Output layer
logits = jnp.dot(h1, w2) + b2
return logits
- Forward Pass: forward performs a forward pass through a two-layer neural network, computing the output (logits) by applying a linear transformation followed by ReLu, and other linear transformations.
Step4: Defining the loss function
A well-defined loss function is essential for guiding the training of our model. In this step, we will implement the mean squared error (MSE) loss function, which measures how well the predicted outputs match the target values, enabling the model to learn effectively.
def loss_fn(params, x, y):
"""Mean squared error loss"""
pred = forward(params, x)
return jnp.mean((pred - y) ** 2)- Loss Function: loss_fn calculates the mean squared error (MSE) loss between the predicted logits and the target labels (y).
Step5: Model Initialization
With our model architecture and loss function defined, we now turn to model initialization. This step involves setting up the parameters of our neural network, ensuring that each layer is ready to begin the training process with random but appropriately scaled weights and biases.
def init_model(rng_key, input_dim, hidden_dim, output_dim):
key1, key2 = jax.random.split(rng_key)
params = [
init_layer_params(key1, input_dim, hidden_dim),
init_layer_params(key2, hidden_dim, output_dim),
]
return params
- Model Initialization: init_model initializes the weights and biases for both layers of the neural networks. It uses two separate random keys for each layer;’s parameter initialization.
Step6: Training Step
Training a neural network involves iterative updates to its parameters based on the computed gradients of the loss function. In this step, we will implement a training function that applies these updates efficiently, allowing our model to learn from the data over multiple epochs.
@jit
def train_step(params, opt_state, x_batch, y_batch):
loss, grads = jax.value_and_grad(loss_fn)(params, x_batch, y_batch)
updates, opt_state = optimizer.update(grads, opt_state)
params = optax.apply_updates(params, updates)
return params, opt_state, loss- Training Step: the train_step function performs a single gradient descent update.
- It calculates the loss and gradients using value_and_grad, which computes both the function values and other gradients.
- The optimizer updates are calculated, and the model parameters are updated accordingly.
- The is JIT-compiled for performance.
Step7: Data and Training Loop
To train our model effectively, we need to generate suitable data and implement a training loop. This section will cover how to create synthetic data for our example and how to manage the training process across multiple batches and epochs.
# Generate some example data
key = PRNGKey(0)
x_data = normal(key, (1000, 10)) # 1000 samples, 10 features
y_data = jnp.sum(x_data**2, axis=1, keepdims=True) # Simple nonlinear function
# Initialize model and optimizer
params = init_model(key, input_dim=10, hidden_dim=32, output_dim=1)
optimizer = optax.adam(learning_rate=0.001)
opt_state = optimizer.init(params)
# Training loop
batch_size = 32
num_epochs = 100
num_batches = x_data.shape[0] // batch_size
# Arrays to store epoch and loss values
epoch_array = []
loss_array = []
for epoch in range(num_epochs):
epoch_loss = 0.0
for batch in range(num_batches):
idx = jax.random.permutation(key, batch_size)
x_batch = x_data[idx]
y_batch = y_data[idx]
params, opt_state, loss = train_step(params, opt_state, x_batch, y_batch)
epoch_loss += loss
# Store the average loss for the epoch
avg_loss = epoch_loss / num_batches
epoch_array.append(epoch)
loss_array.append(avg_loss)
if epoch % 10 == 0:
print(f"Epoch {epoch}, Loss: {avg_loss:.4f}")- Data Generation: Random training data (x_data) and corresponding target (y_data) values are created. Model and Optimizer Initialization: The model parameters and optimizer state are initialized.
- Training Loop: The networks are trained over a specified number of epochs, using mini-batch gradient descent.
- Training loops iterate over batches, performing gradient updates using the train_step function. The average loss per epoch is calculated and stored. It prints the epoch number and the average loss.
Step8: Plotting the Results
Visualizing the training results is key to understanding the performance of our neural network. In this step, we will plot the training loss over epochs to observe how well the model is learning and to identify any potential issues in the training process.
# Plot the results
plt.plot(epoch_array, loss_array, label="Training Loss")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.title("Training Loss over Epochs")
plt.legend()
plt.show()These examples demonstrate how JAX combines high performance with clean, readable code. The functional programming style encouraged by JAX makes it easy to compose operations and apply transformations.
Output:
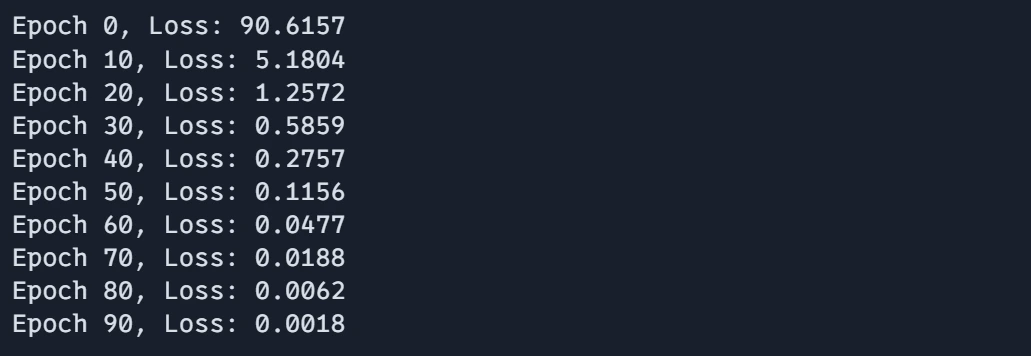
Plot:
These examples demonstrate how JAX combines high performance with clean, readable code. The functional programming style encouraged by JAX makes it easy to compose operations and apply transformations.
Best Practice and Tips
In building neural networks, adhering to best practices can significantly enhance performance and maintainability. This section will discuss various strategies and tips for optimizing your code and improving the overall efficiency of your JAX-based models.
Performance Optimization
Optimizing performance is essential when working with JAX, as it enables us to fully leverage its capabilities. Here, we will explore different techniques for improving the efficiency of our JAX functions, ensuring that our models run as quickly as possible without sacrificing readability.
JIT Compilation Best Practices
Just-In-Time (JIT) compilation is one of the standout features of JAX, enabling faster execution by compiling functions at runtime. This section will outline best practices for effectively using JIT compilation, helping you avoid common pitfalls and maximize the performance of your code.
Bad Function
import jax
import jax.numpy as jnp
from jax import jit
from jax import lax
# BAD: Dynamic Python control flow inside JIT
@jit
def bad_function(x, n):
for i in range(n): # Python loop - will be unrolled
x = x + 1
return x
print("===========================")
# print(bad_function(1, 1000)) # does not work
This function uses a standard Python loop to iterate n times, incrementing the of x by 1 on each iteration. When compiled with jit, JAX unrolls the loop, which can be inefficient, especially for large n. This approach does not fully leverage JAX’s capabilities for performance.
Good Function
# GOOD: Use JAX-native operations
@jit
def good_function(x, n):
return x + n # Vectorized operation
print("===========================")
print(good_function(1, 1000))
This function does the same operation, but it uses a vectorized operation (x+n) instead of a loop. This approach is much more efficient because JAX can better optimize the computation when expressed as a single vectorized operation.
Best Function
# BETTER: Use scan for loops
@jit
def best_function(x, n):
def body_fun(i, val):
return val + 1
return lax.fori_loop(0, n, body_fun, x)
print("===========================")
print(best_function(1, 1000))This approach uses `jax.lax.fori_loop`, which is a JAX-native way to implement loops efficiently. The `lax.fori_loop` performs the same increment operation as the previous function, but it does so using a compiled loop structure. The body_fn function defines the operation for each iteration, and `lax.fori_loop` executes it from o to n. This method is more efficient than unrolling loops and is especially suitable for cases where the number of iterations isn’t known ahead of time.
Output:
===========================
===========================
1001
===========================
1001
The code demonstrates different approaches to handling loops and control flow within JAX’s jit-complied functions.
Memory Management
Efficient memory management is crucial in any computational framework, especially when dealing with large datasets or complex models. This section will discuss common pitfalls in memory allocation and provide strategies for optimizing memory usage in JAX.
Inefficient Memory Management
# BAD: Creating large temporary arrays
@jit
def inefficient_function(x):
temp1 = jnp.power(x, 2) # Temporary array
temp2 = jnp.sin(temp1) # Another temporary
return jnp.sum(temp2)inefficient_function(x): This function creates multiple intermediate arrays, temp1, temp1 and finally the sum of the elements in temp2. Creating these temporary arrays can be inefficient because each step allocates memory and incurs computational overhead, leading to slower execution and higher memory usage.
Efficient Memory Management
# GOOD: Combining operations
@jit
def efficient_function(x):
return jnp.sum(jnp.sin(jnp.power(x, 2))) # Single operationThis version combines all operations into a single line of code. It computes the sine of squared elements of x directly and sums the results. By combining the operation, it avoids creating intermediate arrays, reducing memory footprints and improving performance.
Test Code
x = jnp.array([1, 2, 3])
print(x)
print(inefficient_function(x))
print(efficient_function(x))Output:
[1 2 3]
0.49678695
0.49678695The efficient version leverages JAX’s ability to optimize the computation graph, making the code faster and more memory-efficient by minimizing temporary array creation.
Debugging Strategies
Debugging is an essential part of the development process, especially in complex numerical computations. In this section, we will discuss effective debugging strategies specific to JAX, enabling you to identify and resolve issues quickly.
Using print inside JIT for Debugging
The code shows techniques for debugging within JAX, particularly when using JIT-compiled functions.
import jax.numpy as jnp
from jax import debug
@jit
def debug_function(x):
# Use debug.print instead of print inside JIT
debug.print("Shape of x: {}", x.shape)
y = jnp.sum(x)
debug.print("Sum: {}", y)
return y# For more complex debugging, break out of JIT
def debug_values(x):
print("Input:", x)
result = debug_function(x)
print("Output:", result)
return result
- debug_function(x): This function shows how to use debug.print() for debugging inside a jit compiled function. In JAX, regular Python print statements are not allowed inside JIT due to compilation restrictions, so debug.print() is used instead.
- It prints the shape of the input array x using debug.print()
- After computing the sum of the elements of x, it prints the resulting sum using debug.print()
- Finally, the function returns the computed sum y.
- debug_values(x) function serves as a higher-level debugging approach, breaking out of the JIT context for more complex debugging. It first prints the inputs x using regular print statement. Then calls debug_function(x) to compute the result and finally prints the output before returning the results.
Output:
print("===========================")
print(debug_function(jnp.array([1, 2, 3])))
print("===========================")
print(debug_values(jnp.array([1, 2, 3])))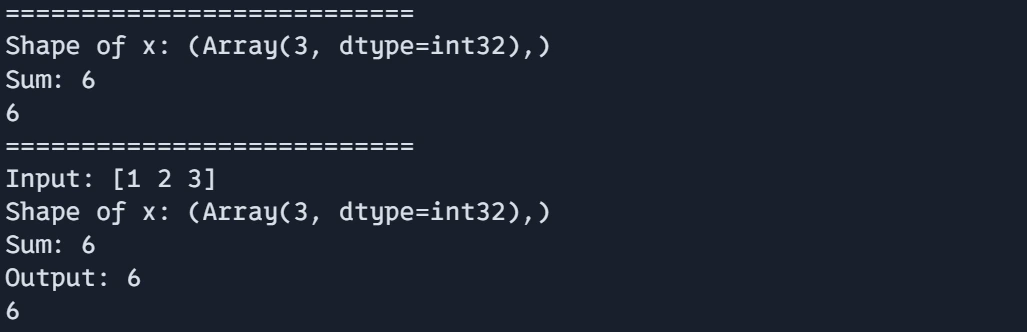
This approach allows for a combination of in-JIT debugging with debug.print() and more detailed debugging outside of JIT using standard Python print statements.
Common Patterns and Idioms in JAX
Finally, we will explore common patterns and idioms in JAX that can help streamline your coding process and improve efficiency. Familiarizing yourself with these practices will aid in developing more robust and performant JAX applications.
Device Memory Management for Processing Large Datasets
# 1. Device Memory Management
def process_large_data(data):
# Process in chunks to manage memory
chunk_size = 100
results = []
for i in range(0, len(data), chunk_size):
chunk = data[i : i + chunk_size]
chunk_result = jit(process_chunk)(chunk)
results.append(chunk_result)
return jnp.concatenate(results)
def process_chunk(chunk):
chunk_temp = jnp.sqrt(chunk)
return chunk_tempThis function processes large datasets in chunks to avoid overwhelming device memory.
It sets chunk_size to 100 and iterates over the data increments of the chunk size, processing each chunk separately.
For each chunk, the function uses jit(process_chunk) to JIT-compile the processing operation, which improves performance by compiling it ahead of time.
The result of each chunk is concatenated into a single array using jnp.concatenated(result) to form a single list.
Output:
print("===========================")
data = jnp.arange(10000)
print(data.shape)
print("===========================")
print(data)
print("===========================")
print(process_large_data(data))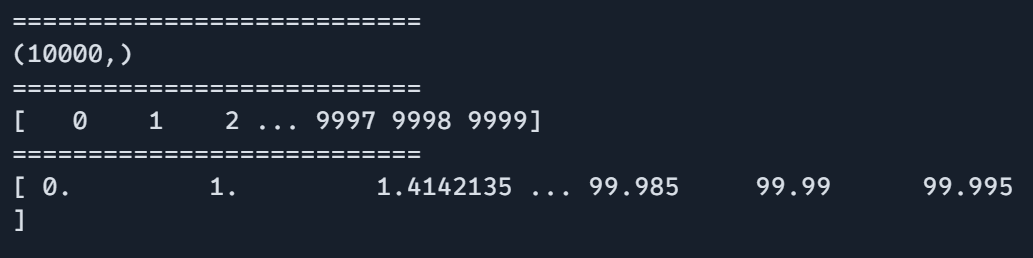
Handling Random Seed for Reproducibility and Better Data Generation
The function create_traing_state() demonstrates managing random number generators (RNGs) in JAX, which is essential for reproducibility and consistent results.
# 2. Handling Random Seeds
def create_training_state(rng):
# Split RNG for different uses
rng, init_rng = jax.random.split(rng)
params = init_network(init_rng)
return params, rng # Return new RNG for next use
It starts with an initial RNG (rng) and splits it into two new RNGs using jax.random.split(). Split RNGs perform different tasks: `init_rng` initializes network parameters, and the updated RNG returns for subsequent operations.
The function returns both the initialized network parameters and the new RNG for further use, ensuring proper handling of random states across different steps.
Now test the code using mock data
def init_network(rng):
# Initialize network parameters
return {
"w1": jax.random.normal(rng, (784, 256)),
"b1": jax.random.normal(rng, (256,)),
"w2": jax.random.normal(rng, (256, 10)),
"b2": jax.random.normal(rng, (10,)),
}
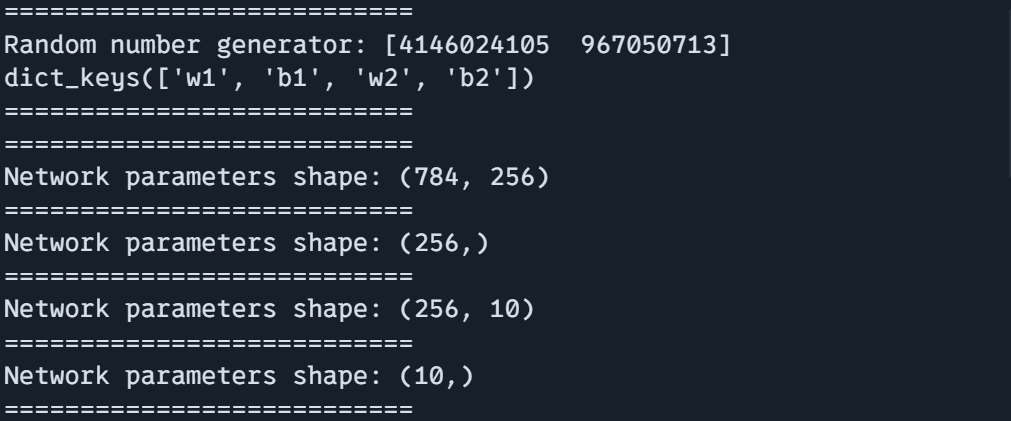
print("===========================")
key = jax.random.PRNGKey(0)
params, rng = create_training_state(key)
print(f"Random number generator: {rng}")
print(params.keys())
print("===========================")
print("===========================")
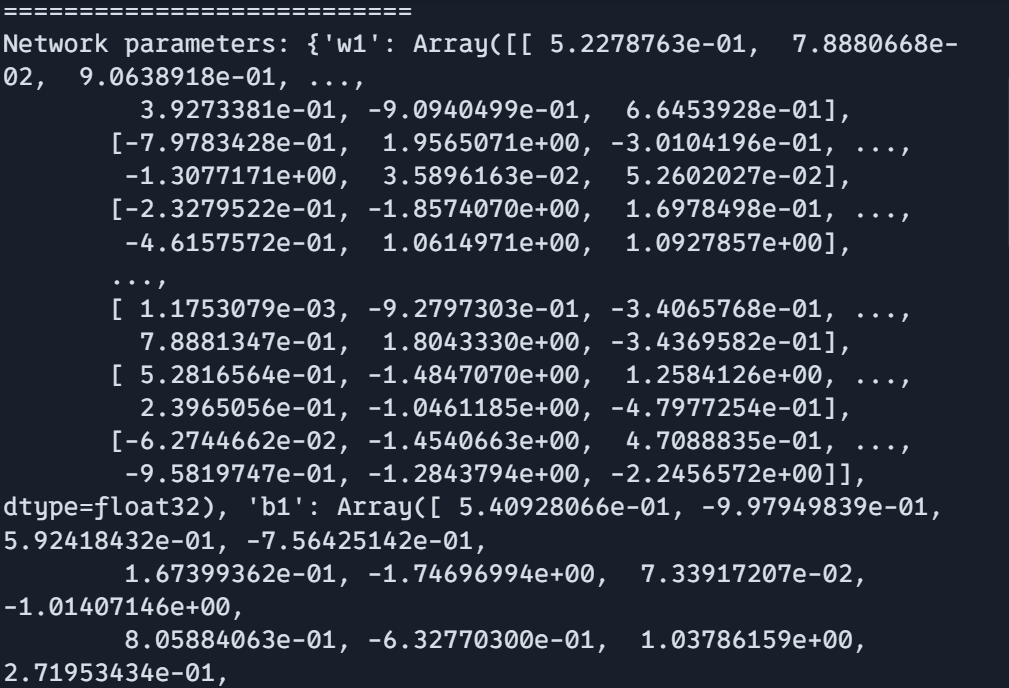
print(f"Network parameters shape: {params['w1'].shape}")
print("===========================")
print(f"Network parameters shape: {params['b1'].shape}")
print("===========================")
print(f"Network parameters shape: {params['w2'].shape}")
print("===========================")
print(f"Network parameters shape: {params['b2'].shape}")
print("===========================")
print(f"Network parameters: {params}")
Output:
Using Static Arguments in JIT
def g(x, n):
i = 0
while i < n:
i += 1
return x + i
g_jit_correct = jax.jit(g, static_argnames=["n"])
print(g_jit_correct(10, 20))Output:
30You can use a static argument if JIT compiles the function with the same arguments each time. This can be useful for the performance optimization of JAX functions.
from functools import partial
@partial(jax.jit, static_argnames=["n"])
def g_jit_decorated(x, n):
i = 0
while i < n:
i += 1
return x + i
print(g_jit_decorated(10, 20))
If You want to use static arguments in JIT as a decorator you can use jit inside of functools. partial() function.
Output:
30Now, we have learned and dived deep into many exciting concepts and tricks in JAX and overall programming style.
What’s Next?
- Experiment with Examples: Try to modify the code examples to learn more about JAX. Build a small project for a better understanding of JAX’s transformations and APIs. Implement classical Machine Learning algorithms with JAX such as Logistic Regression, Support Vector Machine, and more.
- Explore Advanced Topics: Parallel computing with pmap, Custom JAX transformations, Integration with other frameworks
All code used in this article is here
Conclusion
JAX is a powerful tool that provides a wide range of capabilities for machine learning, Deep Learning, and scientific computing. Start with basics, experimenting, and get help from JAX’s beautiful documentation and community. There are so many things to learn and it will not be learned by just reading others’ code you have to do it on your own. So, start creating a small project today in JAX. The key is to Keep Going, learn on the way.
Key Takeaways
- Familiar NumPY-like interface and APIs make learning JAX easy for beginners. Most NumPY code works with minimal modifications.
- JAX encourages clean functional programming patterns that lead to cleaner, more maintainable code and upgradation. But If developers want JAX fully compatible with Object Oriented paradigm.
- What makes JAX’s features so powerful is automatic differentiation and JAX’s JIT compilation, which makes it efficient for large-scale data processing.
- JAX excels in scientific computing, optimization, neural networks, simulation, and machine learning which makes developer easy to use on their respective project.
Frequently Asked Questions
A. Although JAX feels like NumPy, it adds automatic differentiation, JIT compilation, and GPU/TPU support.
A. In a single word big NO, though having a GPU can significantly speed up computation for larger data.
A. Yes, You can use JAX as an alternative to NumPy, though JAX’s APIs look familiar to NumPy JAX is more powerful if you use JAX’s features well.
A. Most NumPy code can be adapted to JAX with minimal changes. Usually just changing import numpy as np to import jax.numpy as jnp.
A. The basics are just as easy as NumPy! Tell me one thing, will you find it hard after reading the above article and hands-on? I answered it for you. YES hard. Every framework, language, libraries is hard not because it is hard by design but because we don’t give much time to explore it. Give it time to get your hand dirty it will be easier day by day.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.





