Following the groundbreaking impact of DeepSeek R1, DeepSeek AI continues to push the boundaries of innovation with its latest offering: Smallpond. This lightweight data processing framework combines the power of DuckDB for SQL analytics and 3FS for high-performance distributed storage, designed to efficiently handle petabyte-scale datasets. Smallpond promises to simplify data processing for AI and big data applications, eliminating the need for long-running services and complex infrastructure, marking another significant leap forward from the DeepSeek team. In this article, we will explore the features, components, and applications of DeepSeek AI’s Smallpond framework, and also learn how to use it.
Learning Objectives
- Learn what DeepSeek Smallpond is and how it extends DuckDB for distributed data processing.
- Understand how to install Smallpond, set up Ray clusters, and configure a computing environment.
- Learn how to ingest, process, and partition data using Smallpond’s API.
- Identify practical use cases like AI training, financial analytics, and log processing.
- Weigh the advantages and challenges of using Smallpond for distributed analytics.
This article was published as a part of the Data Science Blogathon.
Table of Contents
What is DeepSeek Smallpond?
Smallpond is an open-source, lightweight data processing framework developed by DeepSeek AI, designed to extend the capabilities of DuckDB—a high-performance, in-process analytical database—into distributed environments.
By integrating DuckDB with the Fire-Flyer File System (3FS), Smallpond offers a scalable solution for handling petabyte-scale datasets without the overhead of traditional big data frameworks like Apache Spark.
Released on February 28, 2025, as part of DeepSeek’s Open Source Week, Smallpond targets data engineers and scientists who need efficient, simple, and high-performance tools for distributed analytics.
Learn More: DeepSeek Releases 3FS & Smallpond Framework
Key Features of Smallpond
- High Performance: Leverages DuckDB’s native SQL engine and 3FS’s multi-terabyte-per-minute throughput.
- Scalability: Processes petabyte-scale data across distributed nodes with manual partitioning.
- Simplicity: No long-running services or complex dependencies—deploy and use with minimal setup.
- Flexibility: Supports Python (3.8–3.12) and integrates with Ray for parallel processing.
- Open Source: MIT-licensed, fostering community contributions and customization.
Core Components of DeepSeek Smallpond
Now let’s understand the core components of DeepSeek’s Smallpond framework.
DuckDB
DuckDB is an embedded, in-process SQL OLAP database optimized for analytical workloads. It excels at executing complex queries on large datasets with minimal latency, making it ideal for single-node analytics. Smallpond extends DuckDB’s capabilities to distributed systems, retaining its performance benefits.
3FS (Fire-Flyer File System)
3FS is a distributed file system designed by DeepSeek for AI and high-performance computing (HPC) workloads. It leverages modern SSDs and RDMA networking to deliver low-latency, high-throughput storage (e.g., 6.6 TiB/s read throughput in a 180-node cluster). Unlike traditional file systems, 3FS prioritizes random reads over caching, aligning with the needs of AI training and analytics.
Integration of DuckDB and 3FS in Smallpond

Smallpond uses DuckDB as its compute engine and 3FS as its storage backbone. Data is stored in Parquet format on 3FS, partitioned manually by users, and processed in parallel across nodes using DuckDB instances coordinated by Ray. This integration combines DuckDB’s query efficiency with 3FS’s scalable storage, enabling seamless distributed analytics.
Getting Started with Smallpond
Now, let’s learn how to install and use Smallpond.
Step 1: Installation
Smallpond is Python-based and installable via pip available only for Linux distros. Ensure Python 3.8–3.11 is installed, along with a compatible 3FS cluster (or local filesystem for testing).
# Install Smallpond with dependecies
pip install smallpond
# Optional: Install development dependencies (e.g., for testing)
pip install "smallpond[dev]"
# Install Ray Clusters
pip install 'ray[default]'For 3FS, clone and build from the GitHub repository:
git clone https://github.com/deepseek-ai/3fs
cd 3fs
git submodule update --init --recursive
./patches/apply.sh
# Install dependencies (Ubuntu 20.04/22.04 example)
sudo apt install cmake libuv1-dev liblz4-dev libboost-all-dev
# Build 3FS (refer to 3FS docs for detailed instructions)Step 2: Setting Up the Environment
Initialize a ray instance for ray clusters if using 3FS, follow the codes below:
#intialize ray accordingly
ray start --head --num-cpus=<NUM_CPUS> --num-gpus=<NUM_GPUS>Running the above code will produce output similar to the image below:

Now we can initialize Ray with 3FS by using the address we got as shown above. To initialize Ray in smallpond, Configure a compute cluster (e.g., AWS EC2, on-premises) with 3FS deployed on SSD-equipped nodes or For local testing (Linux/Ubuntu), use a filesystem path.
import smallpond
# Initialize Smallpond session (local filesystem for testing)
sp = smallpond.init(data_root="Path/to/local/Storage",ray_address="192.168.214.165:6379")# Enter your own ray address
# For 3FS cluster (update with your 3FS endpoint and ray address)
sp = smallpond.init(data_root="3fs://cluster_endpoint",ray_address="192.168.214.165:6379")# Enter your own ray address
Step 3: Data Ingestion and Preparation
Supported Data Formats
Smallpond primarily supports Parquet files, optimized for columnar storage and DuckDB compatibility. Other formats (e.g., CSV) may be supported via DuckDB’s native capabilities.
Reading and Writing Data
Load and save data using Smallpond’s high-level API.
# Read Parquet file
df = sp.read_parquet("data/input.prices.parquet")
# Process data (example: filter rows)
df = df.map("price > 100") # SQL-like syntax
# Write results back to Parquet
df.write_parquet("data/output/filtered.prices.parquet")Data Partitioning Strategies
Manual partitioning is key to Smallpond’s scalability. Choose a strategy based on your data and workload:
- By File Count: Split into a fixed number of files.
- By Rows: Distribute rows evenly.
- By Hash: Partition based on a column’s hash for balanced distribution.
# Partition by file count
df = df.repartition(3)
# Partition by rows
df = df.repartition(3, by_row=True)
# Partition by column hash (e.g., ticker)
df = df.repartition(3, hash_by="ticker")Step 4: API Referencing
High-Level API Overview
The high-level API simplifies data loading, transformation, and saving:
- read_parquet(path) : Loads Parquet files.
- write_parquet(path) : Saves processed data.
- repartition(n, [by_row, hash_by]) : Partitions data.
- map(expr) : Applies transformations.
Low-Level API Overview
For advanced use, Smallpond integrates DuckDB’s SQL engine and Ray’s task distribution directly:
- Execute raw SQL via partial_sql
- Manage Ray tasks for custom parallelism.
Detailed Function Descriptions
- sp.read_parquet(path): Reads Parquet files into a distributed DataFrame.
df = sp.read_parquet("3fs://data/input/*.parquet")- df.map(expr): Applies SQL-like or Python transformations.
# SQL-like
df = df.map("SELECT ticker, price * 1.1 AS adjusted_price FROM {0}")
# Python function
df = df.map(lambda row: {"adjusted_price": row["price"] * 1.1})- df.partial_sql(query, df): Executes SQL on a DataFrame
df = sp.partial_sql("SELECT ticker, MIN(price), MAX(price) FROM {0} GROUP BY ticker", df)Performance Benchmarks
Smallpond’s performance shines in benchmarks like GraySort, sorting 110.5 TiB across 8,192 partitions in 30 minutes and 14 seconds (3.66 TiB/min throughput) on a 50-node compute cluster with 25 3FS storage nodes.
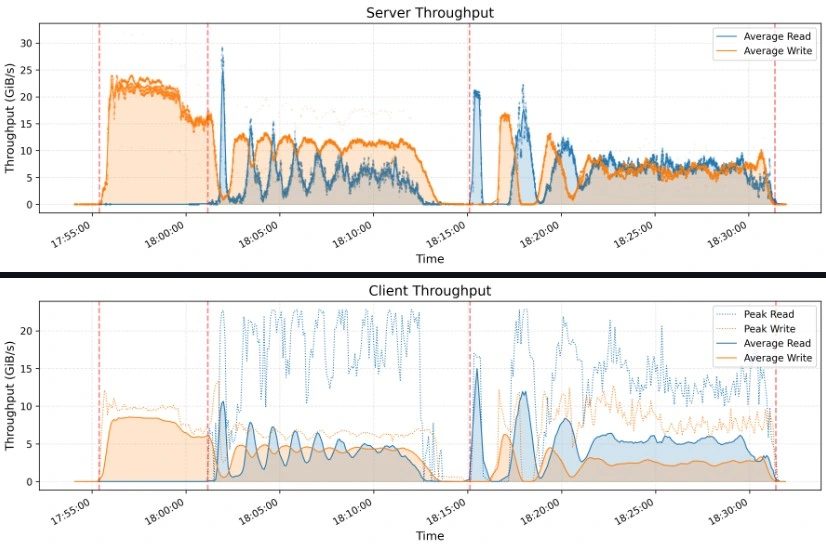
Best Practices for Optimizing Performance
- Partition Wisely: Match partition size to node memory and workload.
- Leverage 3FS: Use SSDs and RDMA for maximum I/O throughput.
- Minimize Shuffling: Pre-partition data to reduce network overhead.
Scalability Considerations
- 10TB–1PB: Ideal for Smallpond with a modest cluster.
- Over 1PB: Requires significant infrastructure (e.g., 180+ nodes).
- Cluster Management: Use managed Ray services (e.g., Anyscale) to simplify scaling.
Applications of Smallpond
- AI Data Pre-processing: Prepare petabyte-scale training datasets.
- Financial Analytics: Aggregate and analyze market data across distributed nodes.
- Log Processing: Process server logs in parallel for real-time insights.
- DeepSeek’s AI Training: Used Smallpond and 3FS to sort 110.5 TiB in under 31 minutes, supporting efficient model training.
Advantages and Disadvantages of Smallpond
| Feature | Advantages | Disadvantages |
|---|---|---|
| Scalability | Handles petabyte-scale data efficiently | Cluster management overhead |
| Performance | Excellent benchmark performance | May not optimize single-node performance |
| Cost | Open-source and cost-effective | Dependence on external frameworks |
| Usability | User-friendly API for ML developers | Security concerns related to DeepSeek’s AI models |
| Architecture | Distributed computing with DuckDB and Ray Core | None |
Conclusion
Smallpond redefines distributed data processing by combining DuckDB’s analytical prowess with 3FS’s high-performance storage. Its simplicity, scalability, and open-source nature make it a compelling choice for modern data workflows. Whether you’re preprocessing AI datasets or analyzing terabytes of logs, Smallpond offers a lightweight yet powerful solution. Dive in, experiment with the code, and join the community to shape its future!
Key Takeaways
- Smallpond is an open-source, distributed data processing framework that extends DuckDB’s SQL capabilities using 3FS and Ray.
- It currently supports only Linux distros and requires Python 3.8–3.12.
- Smallpond is ideal for AI preprocessing, financial analytics, and big data workloads, but requires careful cluster management.
- It is a cost-effective alternative to Apache Spark, with lower overhead and ease of deployment.
- Despite its advantages, it requires infrastructure considerations, such as cluster setup and security concerns with DeepSeek’s models.
The media shown in this article is not owned by Analytics Vidhya and is used at the Author’s discretion.
Frequently Asked Questions
A. DeepSeek Smallpond is an open-source, lightweight data processing framework that extends DuckDB’s capabilities to distributed environments using 3FS for scalable storage and Ray for parallel processing.
A. Smallpond is a lightweight alternative to Spark, offering high-performance distributed analytics without complex dependencies. However, it requires manual partitioning and infrastructure setup, unlike Spark’s built-in resource management.
A. Smallpond requires Python (3.8–3.12), a Linux-based OS, and a compatible 3FS cluster or local storage. For distributed workloads, a Ray cluster with SSD-equipped nodes is recommended.
A. Smallpond primarily supports Parquet files for optimized columnar storage but can handle other formats through DuckDB’s native capabilities.
A. Best practices include manual data partitioning based on workload, leveraging 3FS for high-speed storage, and minimizing data shuffling across nodes to reduce network overhead.
A. Smallpond excels at batch processing but may not be ideal for real-time analytics. For low-latency streaming data, alternative frameworks like Apache Flink or Kafka Streams might be better suited.


